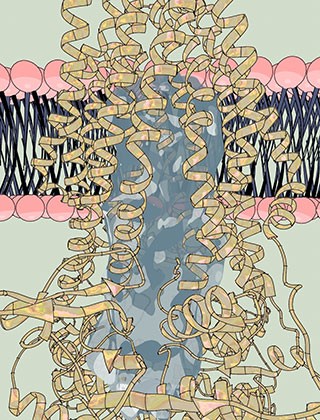
Laboratory of Membrane Biology and Biophysics
A watert passage: The pump, a single-molecule machine, (yellow coils) carries proteins through the cell membrane (pink and dark blue). Within the pump, the researchers found a large water-filled channel (light blue), a natural environment for hydrophilic proteins.
Bacteria have plenty of things to send out into world beyond their own boundaries: coordinating signals to other members of their species, poisons for their enemies, and devious instructions to manipulate host cells they have infected. Before any of this can occur, however, they must first get the shipments past their own cell membranes, and many bacteria have evolved specialized structures and systems for launching the proteins that do these jobs.
Researchers at The Rockefeller University have determined the structure of a simple but previously unexamined pump that controls the passage of proteins through a bacterial cell membrane, an achievement that offers new insight into the mechanics that allow bacteria to manipulate their environments. The results were published in Nature on July 23.
“This pump, called PCAT for peptidase-containing ATP-binding cassette transporter, is composed of a single protein, a sort of all-in-one machine capable of recognizing its cargo, processing it, then burning chemical fuel to pump that cargo out of the cell,” says study author Jue Chen, William E. Ford Professor and head of the Laboratory of Membrane Biology and Biophysics. “This new atomic-level structure explains for the first time the links between these three functions.”
Of the many types of molecules cells need to move into and out of their membranes, proteins are the largest. PCATs specialize in pumping proteins out of the cell, and, because they are single-molecule machines that work alone, or with two partner proteins in some bacteria, they are the simplest such systems.
Each PCAT molecule has three domains, each in duplicate: one recognizes the cargo by a tag it carries, and cuts off that tag; another binds to and burns ATP, a molecule that contains energy stored within its atomic bonds; and the third forms a channel that spans the cells membrane. Previous work had examined the structure of the first two domains, but the structure of the third, had remained a mystery, along with the details of how the components function together.
“At this point, we have no idea how many PCATs exist, although we expect they are numerous, because each specializes in a specific type of cargo. For this study, we focused on one we called PCAT1, which transports a small protein of unknown function,” says first author David Yin-wei Lin, a postdoc in the lab. “To get a sense of how PCAT1 changes shape when powered by energy from ATP, we examined the structure in two states, both with and without ATP.”
The team, which also included Shuo Huang, a research technician who is now a graduate student at Georgia Institute of Technology, purified and crystalized the PCAT1 protein from the heat-loving bacterium Clostridium thermocellum. To determine the structure of the crystals, they used a technique called X-ray diffraction analysis, in which a pattern produced by X-rays bounced off the crystallized protein can be used to infer the structure of the molecule.
The first structure, determined without ATP, revealed a striking feature: a large, water-filled central channel, a natural environment for a water-loving, or hydrophilic, protein. Two side openings into this channel were guarded by the cargo-recognizing domain, acting as a sort of ticket taker. Sites on this domain would recognize and clip off the cargo’s tag, before ushering the protein into the channel.
When ATP is present, they found that the side entrances close, freeing the cargo-recognizing domain to move from its station outside of them. In addition, the ATP-binding domains at the bottom of the channel inside the cell come together. The researchers also saw the water channel shrink, leading them to hypothesize that energy from ATP allows PCAT1 to change conformation in such a way that it pushes its cargo out. This suggests that PCAT1 uses a strategy commonly seen in transport proteins known as alternate access, in which one end of the channel is open while the other closes. However, they qualify that PCATs that transport much larger proteins may function differently.
“By visualizing the structure of this pump, we have been able to determine the details of a transport pathway that, in its simplicity, is fundamentally different from the more complex systems that have been closely studied before. This new information adds to the understanding of how cells send out proteins in order to interact with their environment,” Chen says.
Contact Information
Wynne Parry
Science Writer
wparry@rockefeller.edu
Phone: 212-327-7789