
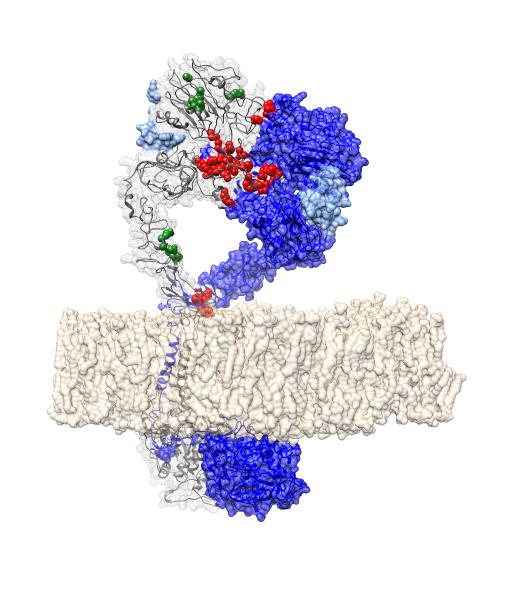
This is a structure showing EGFR -- a cancer driver -- in it's active dimer conformation. Red indicates mutations that destroy the protein-protein interface
Credit: Eduard Porta Pardo
In a collaborative study led by Sanford Burnham Prebys Medical Discovery Institute (SBP), researchers have combined two publicly available 'omics' databases to create a new catalogue of 'cancer drivers'. Cancer drivers are genes that when altered, are responsible for cancer progression.
The researchers used cancer mutation and protein structure databases to identify mutations in patient tumors that alter normal protein-protein interaction (PPI) interfaces. The study, published today in PLoS Computational Biology, identified more than 100 novel cancer driver genes and helps explain how tumors driven by the same gene may lead to different patient outcomes.
“This is the first time that three-dimensional protein features, such as PPIs, have been used to identify driver genes across large cancer datasets,” said lead author Eduard Porta-Pardo, Ph.D., a postdoctoral fellow at SBP.
“We found 71 interfaces in proteins previously unrecognized as cancer drivers, representing potential new cancer predictive markers and/or drug targets. Our analysis also identified several driver interfaces in known cancer genes, such as TP53, HRAS, PI3KCA and EGFR, proving that our method can find relevant cancer driver genes and that alterations in protein interfaces are a common pathogenic mechanism of cancer.”
Cancer is caused by the accumulation of mutations to DNA. Until now, scientists have focused on finding alterations in individual genes and cell pathways that can lead to cancer. But the recent push by the National Institutes of Health (NIH) to encourage data sharing has led to an era of unprecedented ability to systematically analyze large scale genomic, clinical, and molecular data to better explain and predict patient outcomes, as well as finding new drug targets to prevent, treat, and potentially cure cancer.
“For this study we used an extended version of e-Driver, our proprietary computational method of identifying protein regions that drive cancer. We integrated tumor data from almost 6,000 patients in The Cancer Genome Atlas (TCGA) with more than 18,000 three-dimensional protein structures from the Protein Data Bank (PDB),” said Adam Godzik, Ph.D, director of the Bioinformatics and Structural Biology Program at SBP. “The algorithm analyzes whether structural alterations of PPI interfaces are enriched in cancer mutations, and can therefore identify candidate driver genes.”
“Genes are not monolithic black boxes. They have different regions that code for distinct protein domains that are usually responsible for different functions. It's possible that a given protein only acts as a cancer driver when a specific region of the protein is mutated,” Godzik explained. “Our method helps identify novel cancer driver genes and propose molecular hypotheses to explain how tumors apparently driven by the same gene have different behaviors, including patient outcomes.”
“Interestingly, we identified some potential cancer drivers that are involved in the immune system. With the growing appreciation of the importance of the immune system in cancer progression, the immunity genes we identified in this study provide new insight regarding which interactions may be most affected,” Godzik added.
###
The study was performed in collaboration with the European Bioinformatics Institute (UK), Centro de Investigación Principe Felipe (Spain), and CIBER de Enfermedades Raras (Spain).
This study was supported by SPB funds and NIH grant GM101457.
About Sanford Burnham Prebys Medical Discovery Institute
Sanford Burnham Prebys Medical Discovery Institute (SBP) is an independent nonprofit research organization that blends cutting-edge fundamental research with robust drug discovery to address unmet clinical needs in the areas of cancer, neuroscience, immunity, and metabolic disorders. The Institute invests in talent, technology, and partnerships to accelerate the translation of laboratory discoveries that will have the greatest impact on patients. Recognized for its world-class NCI-designated Cancer Center and the Conrad Prebys Center for Chemical Genomics, SBP employs more than 1,100 scientists and staff in San Diego (La Jolla), Calif., and Orlando (Lake Nona), Fla. For more information, visit us at SBPdiscovery.org. The Institute can also be found on Facebook at facebook.com/SBPdiscovery and on Twitter @SBPdiscovery.















