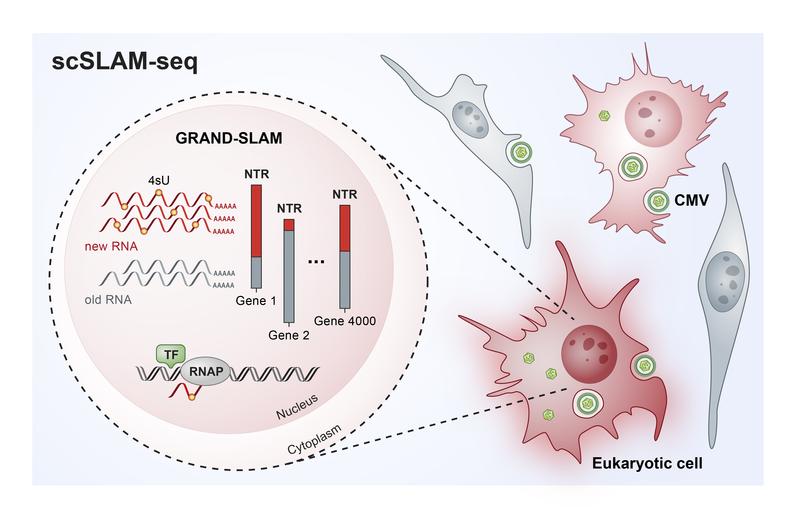
Illustration of the single-cell analysis using scSLAM-seq.
HZI/HIRI/S. Pernitzsch
Cells are equipped with multiple defense mechanisms to fight off intruders. Genes encode the building blocks of the response that must be activated during an enemy attack in an ordered manner. Scientists at the Helmholtz Institute for RNA-based Infection Research (HIRI) in Würzburg, one of the Helmholtz Centre for Infection Research (HZI) sites, and the Julius-Maximilians-Universität Würzburg have developed a new method, scSLAM-seq, which enables them to track the activity of thousands of genes in individual cells over several hours.
For the first time, the researchers were able to explain why some cells are successfully infected by a virus, whereas others are not. In addition, they also gained fundamental new insights into the regulation of genes. Their results are published in the July, 18th issue of the scientific journal Nature.
When viruses enter our bodies – such as during an influenza or a gastrointestinal infection – the processes within the infected cells change: In the worst case, the virus takes the helm and reprograms the cell to its advantage. It then produces viral components on a massive scale allowing the intruder to multiply exponentially.
In other cells, however, the virus may be successfully eliminated by the activation of cellular defense mechanisms. But how can it be that one cell is overrun and another succeeds in getting the virus under control? How quickly do individual cells react to a viral attack and which protective genes are activated?
“Previously, little was known about these issues at the single-cell level,” says Dr. Antoine-Emmanuel Saliba from the Helmholtz Institute for RNA-based Infection Research (HIRI). “But our current study has enabled us to make considerable progress in answering these questions.”
Prof. Florian Erhard, from the Institute for Virology and Immunobiology at the Julius-Maximilians-Universität (JMU) in Würzburg, Saliba from the HIRI and Prof. Lars Dölken, who works at both JMU and HIRI, teamed up in investigating how gene activity – which reflects the identity and the physiological state of a cell – changes in individual infected mouse cells after an infection with cytomegalovirus.
Cytomegaloviruses are widespread, with more than 80 percent of people infected. Generally, healthy people are not harmed by the virus, however an infection in newborns or transplant patients can become dangerous and have serious neurological consequences. An experimental method, single-cell RNA sequencing (scRNAseq), that emerged only a few years ago, can be used to determine which genes of a cell are currently active.
However, short-term changes in gene activity, such as those that occur during a viral infection, can hardly be detected. In addition, each individual cell can only be examined once. With current technologies it therefore remained unclear, how individual cells react to external stimuli, for example a viral infection.
In order to investigate the molecular processes taking place in individual infected cells, the researchers developed a new method, called scSLAM-seq, which, for the first time, enables then to visualize which genes are activated in individual cells within the time frame of a few hours. When a gene is activated, an RNA (ribonucleic acid) copy of the DNA is produced.
RNA consists of four different components, namely the organic bases adenine, cytosine, guanine and uracil. These are arranged in a chain and in a combination that is characteristic of the original gene. This serves as a template for the production of a protein that performs a specific function within the cell.
scSLAM-seq sorts RNA into old and new
In order to distinguish between RNA that was already present before viral infection and new RNA that was made thereafter, the researchers utilized a labelling trick: They added a form of the RNA component uracil, which was chemically modified in comparison to its natural variant, to the culture medium of the cells at the same time as the infecting virus. The cells then incorporated the labelled uracil into their newly produced RNA.
After extracting RNA from cells two hours later, the labelled uracil was converted into another RNA component, cytosine, via a chemical reaction. “The RNA sequence contains cytosine whereas uracil actually should have been incorporated,” explains Dölken. “The idea behind this is that the RNA produced after infection now has a label that can be used to identify it as ‘new’ during subsequent RNA sequencing.”
Using a novel patented bioinformatic method, the researchers investigated the RNA of each individual cell. In this way, they were able to commonly assess the expression of more than 4000 genes per cell and precisely quantify the relative contributions of new and old RNAs. “The data that we can collect using scSLAM-seq are spectacular,” says Erhard. “For each individual cell we can determine what percentage of its RNA is newly produced within two hours after a challenge – in this case, a viral infection.
For the first time, this enables dose-response analyses at the single-cell level.” The research team investigated the RNA of 100 single cells in total. “This was sufficient to obtain an entirely new insight into cellular gene activation,” says Saliba. “For the first time, scSLAM-seq enables us, to precisely analyze how a single cell reacts to a disruption such as a viral or bacterial infection within a short timeframe and which genes are subsequently read more or less often, enabling us to understand which battle plan the cell has prepared for the fight against an intruder.”
Each cell works differently
The scientists were also able to show that the production of RNA from a gene, called transcription, is not continuous, but takes place in bursts: During viral infection hundreds of genes that are otherwise transcriptionally silent are activated. In particular, this includes genes that help our cells to fight the infection.
“In our analyses, many cellular genes exhibited pronounced burst behaviour. Within a cell, almost the entire RNA of a gene was either new or old,” says Erhard, “and we were able to show that the burst behaviour depends on the structure of certain regions of the gene sequence – the promoters – which control the transcription of genes.”
Transcription is therefore not just stronger or weaker in infected cells, but occurs according to an on/off principle. Saliba: “This also explains why cells often have such different RNA profiles and why some cells are able to fight viruses immediately upon first contact and why others first require help from neighbouring cells.”
Each cell works according to its own rhythm: Cells with initially identical RNA profiles will display completely different RNAs after only a few days. Dölken: “Our investigations enabled us to gain groundbreaking insights into the timing of cellular gene activation, which have in fact completely changed my understanding of this area.”
Protection against autoimmune diseases
The on/off principle of cellular gene activation likely has a very important function for us: If all genes used to fight viruses were continuously produced by every cell in the body, this could lead to unwanted reactions and autoimmune diseases. “By switching on at the right moment – but only when absolutely necessary – our immune system can create a protective cellular environment without risk of harmful false reactions that may lead to autoimmunity,” says Dölken. For this reason, certain mechanisms are fully functional in only a small number of the body's cells.
These “sentinel” cells are then able to detect an invading virus and fight it efficiently, for example. They inform the other cells, which then also start up the complete defence arsenal and activate the appropriate genes in order to control the infection and avert danger.
Using their patented bioinformatics analysis method named “GRAND-SLAM”, which is necessary for evaluating the highly complex experimental data, the researchers in Würzburg obtained new insights into hidden cellular processes. “For the first time, we can really see how a single cell reacts,” says Dölken. It can be used in future investigations for answering a wide range of questions at the single-cell level. “scSLAM-seq is ideal for knock-out studies in order to discover which genes play a key role in fighting pathogens or in the development of diseases,” says Saliba. “It is methodologically simple, the data quality is high and ideal for dose-response studies, also over time.” scSLAM-seq and GRAND-SLAM will thus have major implications in other fields of natural science like cancer and drug development.
The Helmholtz Centre for Infection Research:
Scientists at the Helmholtz Centre for Infection Research (HZI) in Braunschweig, Germany, are engaged in the study of different mechanisms of infection and of the body’s response to infection. Helping to improve the scientific community’s understanding of a given bacterium’s or virus’ pathogenicity is key to developing effective new treatments and vaccines. The HZI is a member of the German Center for Infection Research (DZIF). http://www.helmholtz-hzi.de/en
The Helmholtz Institute for RNA-based Infection Research:
The Helmholtz Institute for RNA-based Infection Research (HIRI) was founded in May 2017. It is a joint institution of the Helmholtz Centre for Infection Research (HZI) in Braunschweig and the Julius-Maximilians-Universität Würzburg (JMU). Based on the University hospital campus, the HIRI is the first federal institute to focus on the role of ribonucleic acids (RNAs) in infection processes. Its mission is to combine basic research with the development of new RNA-centric therapeutic approaches to treat infections. http://www.helmholtz-hiri.de
Contact:
Susanne Thiele, Press Officer
susanne.thiele@helmholtz-hzi.de
Dr Andreas Fischer, Editor
andreas.fischer@helmholtz-hzi.de
Helmholtz Centre for Infection Research
Press and Communications
Inhoffenstr. 7
D-38124 Braunschweig
Germany
Phone: +49 531 6181-1404
Florian Erhard, Marisa A.P. Baptista, Tobias Krammer, Thomas Hennig, Marius Lange, Panagiota Arampatzi, Christopher Jürges, Fabian J. Theis, Antoine-Emmanuel Saliba, Lars Dölken: scSLAM-seq reveals core features of transcription dynamics in single cells. Nature 2019, doi: 10.1038/s41586-019-1369-y