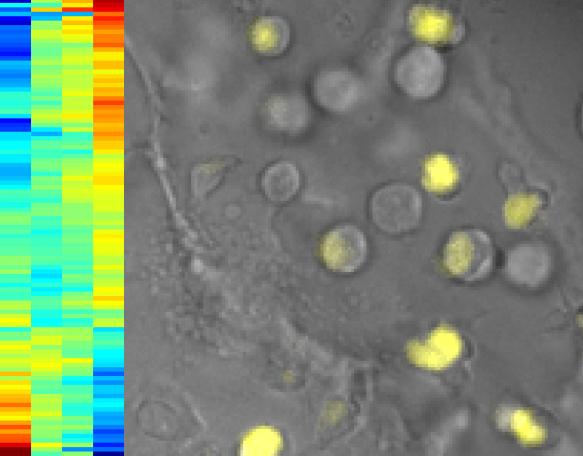
Researchers imaged cells to identify proteins that affect the expression of a genetic switch for T cells. On the right, T cells where the switch is activated glow in yellow. On the left, the rainbow pattern, a hierarchical cluster analysis, tells researchers which genes are controlled by the switch. The horizontal stripes are the genes. If they stripes turn red going from left to right, it means they are turning on; if they turn blue, the genes are turning off.
Credit: Caltech
“We already know which genetic switch directs cells to commit to becoming T cells, but we wanted to figure out what enables that switch to be turned on,” says Hao Yuan Kueh, a postdoctoral scholar at Caltech and lead author of a Nature Immunology report about the work, published on July 4.
The study found that a group of four proteins, specifically DNA-binding proteins known as transcription factors, work in a multi-tiered fashion to control the T-cell genetic switch in a series of steps. This was a surprise because transcription factors are widely assumed to work in a simultaneous, all-at-once fashion when collaborating to regulate genes.
The results may ultimately allow doctors to boost a person's T-cell population. This has potential applications in fighting various diseases, including AIDS, which infects mature T cells.
“In the past, combinatorial gene regulation was thought to involve all the transcription factors being required at the same time,” says Kueh, who works in the lab of Ellen Rothenberg, Caltech's Albert Billings Ruddock Professor of Biology.
“This was particularly true in the case of the genetic switch for T-cell commitment, where it was thought that a quorum of the factors working simultaneously was needed to ensure that the gene would only be expressed in the right cell type.”
The authors report that a key to their finding was the ability to image live cells in real-time. They genetically engineered mouse cells so that a gene called Bcl11b–the key switch for T cells–would express a fluorescent protein in addition to its own Bcl11b protein. This caused the mouse cells to glow when the Bcl11b gene was turn on. By monitoring how different transcription factors, or proteins, affected the activation of this genetic switch in individual cells, the researchers were able to isolate the distinct roles of the proteins.
The results showed that four proteins work together in three distinct steps to flip the switch for T cells. Kueh says to think of the process as a team of people working together to get a light turned on. He says first two proteins in the chain (TCF1 and GATA3) open a door where the main light switch is housed, while the next protein (Notch) essentially switches the light on. A fourth protein (Runx1) controls the amplitude of the signal, like sliding a light dimmer.
“We identify the contributions of four regulators of Bcl11b, which are all needed for its activation but carry out surprisingly different functions in enabling the gene to be turned on,” says Rothenberg. “It's interesting–the gene still needs the full quorum of transcription factors, but we now find that it also needs them to work in the right order. This makes the gene respond not only to the cell's current state, but also to the cell's recent developmental history.”
Team member Kenneth Ng, a visiting student from California Polytechnic State University, says he was surprised by how much detail they could learn about gene regulation using live imaging of cells.
“I had read about this process in textbooks, but here in this study we could pinpoint what the proteins are really doing,” he says.
The next step in the research is to get a closer look at precisely how the T cell genetic switch itself works. Kueh says he wants to “unscrew the panels” of the switch and understand what is physically going on in the chromosomal material around the Bcl11b gene.
###
The Nature Immunology paper, titled, “Asynchronous combinatorial action of four regulatory factors activates Bcl11b for T cell commitment,” includes seven additional Caltech coauthors: Mary Yui, Shirley Pease, Jingli Zhang, Sagar Damle, George Freedman, Sharmayne Siu, and Michael Elowitz; as well as a collaborator at the Fred Hutchinson Cancer Research Center, Irwin Bernstein. The work at Caltech was funded by a CRI/Irvington Postdoctoral Fellowship, the National Institutes of Health, the California Institute for Regenerative Medicine, the Al Sherman Foundation, and the Louis A. Garfinkle Memorial Laboratory Fund.