Making nanowires from protein and DNA
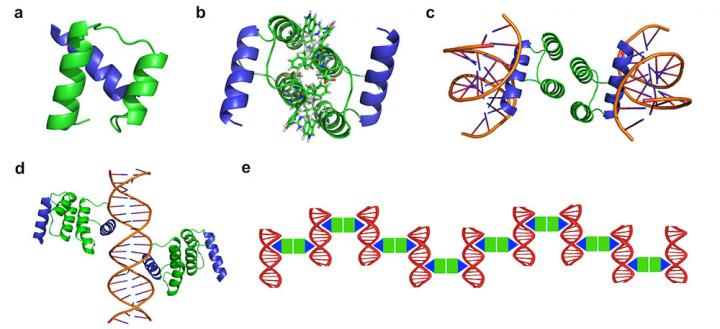
Design strategy of protein-DNA nanowires. The protein-DNA nanowire is self-assembled with a computationally designed protein homodimer and a double-stranded DNA with the protein binding sites properly arranged. Credit: Yun (Kurt) Mou, Jiun-Yann Yu, Timothy M. Wannier, Chin-Lin Guo and Stephen L. Mayo/Caltech
For example, synthetic structures made of DNA could one day be used to deliver cancer drugs directly to tumor cells, and customized proteins could be designed to specifically attack a certain kind of virus.
Although researchers have already made such structures out of DNA or protein alone, a Caltech team recently created–for the first time–a synthetic structure made of both protein and DNA. Combining the two molecule types into one biomaterial opens the door to numerous applications.
A paper describing the so-called hybridized, or multiple component, materials appears in the September 2 issue of the journal Nature.
There are many advantages to multiple component materials, says Yun (Kurt) Mou (PhD '15), first author of the Nature study. “If your material is made up of several different kinds of components, it can have more functionality. For example, protein is very versatile; it can be used for many things, such as protein-protein interactions or as an enzyme to speed up a reaction. And DNA is easily programmed into nanostructures of a variety of sizes and shapes.”
But how do you begin to create something like a protein-DNA nanowire–a material that no one has seen before?
Mou and his colleagues in the laboratory of Stephen Mayo, Bren Professor of Biology and Chemistry and the William K. Bowes Jr. Leadership Chair of Caltech's Division of Biology and Biological Engineering, began with a computer program to design the type of protein and DNA that would work best as part of their hybrid material.
“Materials can be formed using just a trial-and-error method of combining things to see what results, but it's better and more efficient if you can first predict what the structure is like and then design a protein to form that kind of material,” he says.
The researchers entered the properties of the protein-DNA nanowire they wanted into a computer program developed in the lab; the program then generated a sequence of amino acids (protein building blocks) and nitrogenous bases (DNA building blocks) that would produce the desired material.
However, successfully making a hybrid material is not as simple as just plugging some properties into a computer program, Mou says. Although the computer model provides a sequence, the researcher must thoroughly check the model to be sure that the sequence produced makes sense; if not, the researcher must provide the computer with information that can be used to correct the model. “So in the end, you choose the sequence that you and the computer both agree on. Then, you can physically mix the prescribed amino acids and DNA bases to form the nanowire.”
The resulting sequence was an artificial version of a protein-DNA coupling that occurs in nature. In the initial stage of gene expression, called transcription, a sequence of DNA is first converted into RNA. To pull in the enzyme that actually transcribes the DNA into RNA, proteins called transcription factors must first bind certain regions of the DNA sequence called protein-binding domains.
Using the computer program, the researchers engineered a sequence of DNA that contained many of these protein-binding domains at regular intervals. They then selected the transcription factor that naturally binds to this particular protein-binding site–the transcription factor called Engrailed from the fruit fly Drosophila. However, in nature, Engrailed only attaches itself to the protein-binding site on the DNA. To create a long nanowire made of a continuous strand of protein attached to a continuous strand of DNA, the researchers had to modify the transcription factor to include a site that would allow Engrailed also to bind to the next protein in line.
“Essentially, it's like giving this protein two hands instead of just one,” Mou explains. “The hand that holds the DNA is easy because it is provided by nature, but the other hand needs to be added there to hold onto another protein.”
Another unique attribute of this new protein-DNA nanowire is that it employs coassembly–meaning that the material will not form until both the protein components and the DNA components have been added to the solution. Although materials previously could be made out of DNA with protein added later, the use of coassembly to make the hybrid material was a first. This attribute is important for the material's future use in medicine or industry, Mou says, as the two sets of components can be provided separately and then combined to make the nanowire whenever and wherever it is needed.
This finding builds on earlier work in the Mayo lab, which, in 1997, created one of the first artificial proteins, thus launching the field of computational protein design. The ability to create synthetic proteins allows researchers to develop proteins with new capabilities and functions, such as therapeutic proteins that target cancer. The creation of a coassembled protein-DNA nanowire is another milestone in this field.
“Our earlier work focused primarily on designing soluble, protein-only systems. The work reported here represents a significant expansion of our activities into the realm of nanoscale mixed biomaterials,” Mayo says.
Although the development of this new biomaterial is in the very early stages, the method, Mou says, has many promising applications that could change research and clinical practices in the future.
“Our next step will be to explore the many potential applications of our new biomaterial,” Mou says. “It could be incorporated into methods to deliver drugs into cells–to create targeted therapies that only bind to a certain biomarker on a certain cell type, such as cancer cells. We could also expand the idea of protein-DNA nanowires to protein-RNA nanowires that could be used for gene therapy applications. And because this material is brand-new, there are probably many more applications that we haven't even considered yet.”
###
The work was published in a paper titled, “Computational design of co-assembling protein-DNA nanowires.” In addition to Mou and Mayo, other Caltech coauthors include former graduate students Jiun-Yann Yu (PhD '14) and Timothy M. Wannier (PhD '15), as well as Chin-Lin Guo from Academia Sinica in Taiwan. The work was funded by the Defense Advanced Research Projects Agency Protein Design Processes Program, a National Security Science and Engineering Faculty Fellowship, and the Caltech Programmable Molecular Technology Initiative funded by the Gordon and Betty Moore Foundation.
Media Contact
All latest news from the category: Life Sciences and Chemistry
Articles and reports from the Life Sciences and chemistry area deal with applied and basic research into modern biology, chemistry and human medicine.
Valuable information can be found on a range of life sciences fields including bacteriology, biochemistry, bionics, bioinformatics, biophysics, biotechnology, genetics, geobotany, human biology, marine biology, microbiology, molecular biology, cellular biology, zoology, bioinorganic chemistry, microchemistry and environmental chemistry.
Newest articles

NASA: Mystery of life’s handedness deepens
The mystery of why life uses molecules with specific orientations has deepened with a NASA-funded discovery that RNA — a key molecule thought to have potentially held the instructions for…

What are the effects of historic lithium mining on water quality?
Study reveals low levels of common contaminants but high levels of other elements in waters associated with an abandoned lithium mine. Lithium ore and mining waste from a historic lithium…

Quantum-inspired design boosts efficiency of heat-to-electricity conversion
Rice engineers take unconventional route to improving thermophotovoltaic systems. Researchers at Rice University have found a new way to improve a key element of thermophotovoltaic (TPV) systems, which convert heat…



