Carnegie Mellon develops new method for analyzing synaptic density
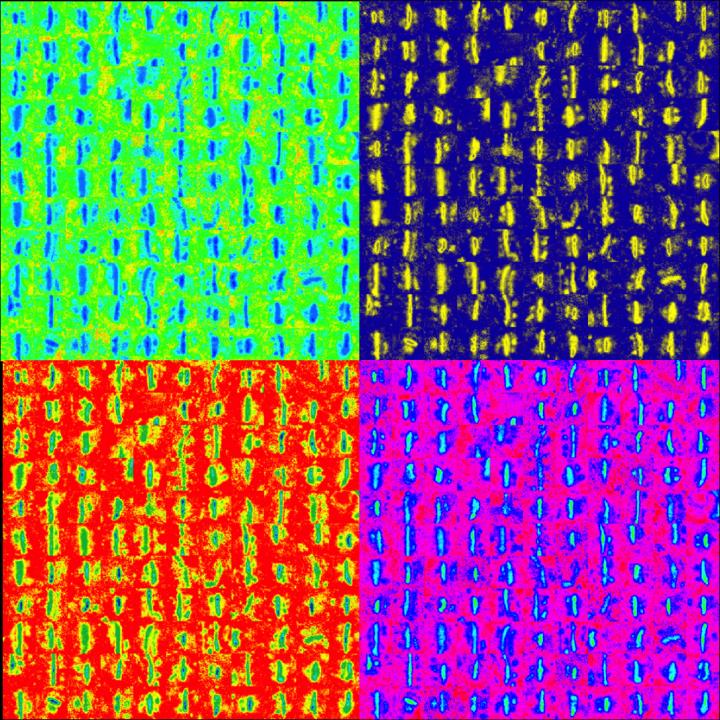
This image shows synapses in the somatosensory cortex stained with ethanolic phosphotungstic acid and visualized using electron microscopy. Synapses were identified using a Carnegie Mellon-developed machine learning algorithm that enables a high-throughput analysis of experience-dependent changes in synapse properties across the cortical column. For this image, candidate synapses were selected from electron micrographs and aligned, then pseudocolored for contrast. Image credit: Saket Navlakha and Alison Barth; the Journal of Neuroscience.
Carnegie Mellon University researchers have developed a new approach to broadly survey learning-related changes in synapse properties. In a study published in the Journal of Neuroscience and featured on the journal's cover, the researchers used machine-learning algorithms to analyze thousands of images from the cerebral cortex.
This allowed them to identify synapses from an entire cortical region, revealing unanticipated information about how synaptic properties change during development and learning. The study is one of the largest electron microscopy studies ever carried out, evaluating more subjects and more images than prior researchers have attempted.
As the brain learns and responds to sensory stimuli, its neurons make connections with one another. These connections, called synapses, facilitate neuronal communication, and their anatomic and electrophysiological properties contain information vital to understanding how the brain behaves in health and disease. Researchers use different techniques, including electron microscopy, to identify and analyze synapse properties. While electron microscopy can be a useful tool for reconstructing neural circuits, it is also data and labor intensive. As a result, researchers have only been able to use it to study small, targeted areas of the brain until now.
Studying a large section of the brain using traditional electron microscopy techniques would result in terabytes of unwieldy data, given that the brain has billions of neurons, each with hundreds to thousands of synaptic connections. The new technique developed at Carnegie Mellon simplifies this problem by combining a specialized staining process with machine learning.
“Instead of getting perfect information from a tiny part of the brain, we can now get lower-resolution information from a huge region of the brain,” said Alison Barth, professor of biological sciences and interim director of Carnegie Mellon's BrainHub neuroscience initiative. “This could be a great tool to see how disease progresses, or how drug treatments alter or restore synaptic connections.”
This research is the latest example of how researchers with Carnegie Mellon's BrainHub research initiative are combining their expertise in biology and computer science to create new tools to advance neuroscience. The technique uses a special chemical preparation that deeply stains the synapses in a sample of brain tissue. When the tissue is imaged using an electron microscope, only the synapses can be seen, creating an image that can be easily classified by a computer program. Researchers then use machine learning algorithms to identify and compare synapse properties across a column of the cerebral cortex.
To test the effectiveness of their technique, the researchers, led by Santosh Chandrasekaran, examined how synapses across a complex circuit, composed of hundreds of interconnected neurons, would change with altered somatosensory input. In the past, Barth has used this model to study how neurons behave and synapses form in both learning and development. But traditional techniques only allowed her to look at neurons in a very small area of the neocortex.
“It was like looking for the perfect gift, but only going to one store. We might have been able to find something at that first location, but it was always possible that we might find something else – maybe even something better – at another place,” said Barth, who is a member of the joint Carnegie Mellon/University of Pittsburgh Center for the Neural Basis of Cognition (CNBC). “This new technique allows us to look across all six layers of the neocortex, and to see how synapses across different parts of the circuit change together.”
The researchers analyzed close to 25,000 images and 40,000 synapses, exponentially more than they were ever able to look at before using traditional methods. They found that the technique could be used to determine increases in synapse density and size during development and learning. Most notably, they found that synapse properties changed in a coordinated way across the entire region of the neocortex examined.
“Some of the cortical layers we saw were most affected have never been examined systematically before,” explains Barth. “We've got a lot of great leads to follow up on.”
The researchers are now beginning to use this data to develop new hypotheses about how synapses are organized in the neocortex in response to sensory input.
###
Additional study authors include: Saket Navlakha, formerly of Carnegie Mellon and now at the Salk Institute for Biological Studies; Nicholas J. Audette, Dylan D. McCreary, and Joe Suhan of Carnegie Mellon's Department of Biological Sciences and the CNBC; and Ziv Bar-Joseph of Carnegie Mellon's Machine Learning Department and Lane Center for Computational Cancer Research.
This research was funded by the National Institutes of Health (DA017188, MH099784), the McKnight Foundation, the Society for Neuroscience, the National Science Foundation (135-6505) and the James S. McDonnell Foundation.
As the birthplace of artificial intelligence and cognitive psychology, Carnegie Mellon has been a leader in the study of brain and behavior for more than 50 years. The university has created some of the first cognitive tutors, helped to develop the Jeopardy-winning Watson, founded a groundbreaking doctoral program in neural computation, and completed cutting-edge work in understanding the genetics of autism. Building on its strengths in biology, computer science, psychology, statistics and engineering, CMU recently launched BrainHub, a global initiative that focuses on how the structure and activity of the brain give rise to complex behaviors.
Media Contact
All latest news from the category: Life Sciences and Chemistry
Articles and reports from the Life Sciences and chemistry area deal with applied and basic research into modern biology, chemistry and human medicine.
Valuable information can be found on a range of life sciences fields including bacteriology, biochemistry, bionics, bioinformatics, biophysics, biotechnology, genetics, geobotany, human biology, marine biology, microbiology, molecular biology, cellular biology, zoology, bioinorganic chemistry, microchemistry and environmental chemistry.
Newest articles

A ‘language’ for ML models to predict nanopore properties
A large number of 2D materials like graphene can have nanopores – small holes formed by missing atoms through which foreign substances can pass. The properties of these nanopores dictate many…

Clinically validated, wearable ultrasound patch
… for continuous blood pressure monitoring. A team of researchers at the University of California San Diego has developed a new and improved wearable ultrasound patch for continuous and noninvasive…

A new puzzle piece for string theory research
Dr. Ksenia Fedosova from the Cluster of Excellence Mathematics Münster, along with an international research team, has proven a conjecture in string theory that physicists had proposed regarding certain equations….



