Resurrecting ancient protein partners reveals origin of protein regulation
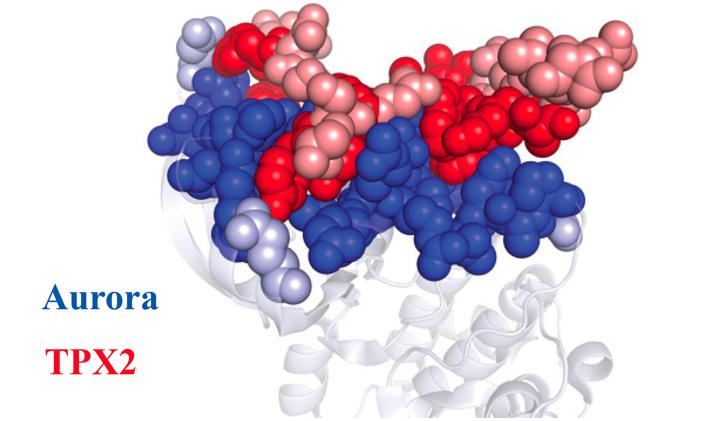
Once the Aurora A protein (blue) and TPX2 protein (red) learned how to fit together, they stayed that way forever. Dark dots represent amino acids that remain unchanged throughout evolution. Light dots represent amino acids that differ between the ancient and modern proteins. Credit: Dorothee Kern/Brandeis University/HHMI
Biochemist Dorothee Kern and her colleagues reconstructed extinct forms of two cell division proteins, and then watched in the lab as one boosted the activity of the other. What Kern's team observed represents the earliest known instance of two proteins interacting in such a way, the researchers reported February 21 in the journal Science.
That interaction, called allosteric regulation, occurs between many modern proteins, but “how such a feature evolved is unexplored territory,” Kern says.
Over the eons, allosteric regulation has become an indispensable tool for cells. As cells developed more elaborate networks of protein activity, Kern explains, they needed more nuanced controls to tweak them.
“Allosteric regulation is arguably the biggest evolutionary step in the development of higher organisms,” she says.
Kern, a Howard Hughes Medical Institute (HHMI) Investigator at Brandeis University, wants to unravel how proteins and other molecules work together – at atomic resolution and in real time. Pinpointing how proteins behave as they send cellular signals, bind to other molecules, or kickstart chemical reactions could give scientists a new window into drug discovery.
Kern chose to study a protein called Aurora A because it's vitally important in cell division – abnormalities can drive cancer growth – and because it's turned on by a protein partner called TPX2.
During cell division, Aurora A helps to distribute chromosomes equally between daughter cells. TPX2 binds to an “allosteric site” of Aurora A – a spot apart from the active site of the protein. Then, TPX2 brings Aurora A to the site of the action and switches the protein into high gear. Kern wanted to know how the two proteins' partnership evolved.
Organisms across the evolutionary tree make some version of the Aurora A and TPX2 proteins. First, Kern, study coauthor Adelajda Hadzipasic, and their colleagues at Brandeis compared versions of Aurora A and TPX2 from a diverse set of species – including bacteria, plants, and humans. Then, they did a bioinformatics analysis, traveling back in time to deduce the ancient amino acid sequences of the two proteins' oldest common ancestors. Next, they used those sequences to recreate the ancient proteins.
“Evolution used to be speculative subject, but it is now experimental. That's what's really exciting about this technique,” says Christopher Miller, an HHMI Investigator alum and recently retired biochemist who used to work next door to Kern at Brandeis.
Experiments in the lab let Kern's team map the proteins' entwined histories. Initially, around 1.5 billion years ago, Aurora A had to self-activate – no TPX2 required. After TPX2 showed up, about a half billion years later, the protein increased Aurora A's productivity by pulling it to the job site. “That created an evolutionary increase in fitness,” she says.
Because teaming up with TPX2 boosted Aurora A's efficiency, the partnership endured. That gave the two proteins time to dial in a more sophisticated relationship, Kern says. Over time, TPX2 took on an additional, new role – as Aurora A's activator. Today, the modern Aurora A/TPX2 pair can work 10 times faster than Aurora A alone.
And when Kern's team combined ancient Aurora A with modern TPX2, they found that the two proteins bound together surprisingly well – despite almost a billion years of evolution between them.
That wasn't expected, Kern says. Conventional wisdom held that proteins that work together also evolve together – so a change in one partner is matched by a complementary change in the other. Modern TPX2, the researchers thought, would likely have amassed too many changes to connect with such an ancient partner.
Kern solved this mystery. The sections of Aurora A and TPX2 that touch stayed essentially the same over 1 billion years, her team discovered, even as other parts of the proteins morphed and shifted over time.
Investigating the details of such interactions could provide a springboard for creating new types of drugs, says Neel Shah, a chemist at Columbia University. Though there's a long road between identifying proteins' allosteric sites and actually designing drugs that can take advantage of them, he says, “these findings could be a starting point.”
###
Citation
Adelajda Hadzipasic et al. “Ancient origins of allosteric activation in a Ser-Thr kinase.” Science. Published February 21, 2020. doi: 10.1126/science.aay9959
Media Contact
All latest news from the category: Life Sciences and Chemistry
Articles and reports from the Life Sciences and chemistry area deal with applied and basic research into modern biology, chemistry and human medicine.
Valuable information can be found on a range of life sciences fields including bacteriology, biochemistry, bionics, bioinformatics, biophysics, biotechnology, genetics, geobotany, human biology, marine biology, microbiology, molecular biology, cellular biology, zoology, bioinorganic chemistry, microchemistry and environmental chemistry.
Newest articles

NASA: Mystery of life’s handedness deepens
The mystery of why life uses molecules with specific orientations has deepened with a NASA-funded discovery that RNA — a key molecule thought to have potentially held the instructions for…

What are the effects of historic lithium mining on water quality?
Study reveals low levels of common contaminants but high levels of other elements in waters associated with an abandoned lithium mine. Lithium ore and mining waste from a historic lithium…

Quantum-inspired design boosts efficiency of heat-to-electricity conversion
Rice engineers take unconventional route to improving thermophotovoltaic systems. Researchers at Rice University have found a new way to improve a key element of thermophotovoltaic (TPV) systems, which convert heat…



