A taste for plastic
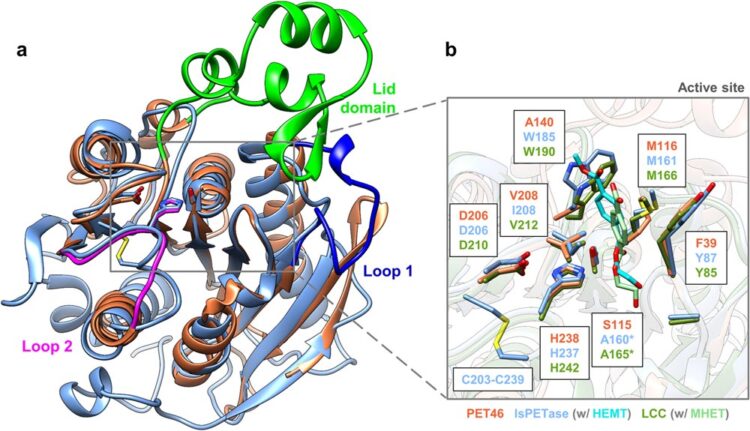
The structure of PET46 is similar to that of known enzymes such as IsPETase and LCC, but has unique features. One example is the unusual 'lid', shown in light green.
© Perez-Garcia, P., Chow, J., Costanzi, E. et al. (2023).
Discovered for the first time: New deep-sea enzyme breaks down PET plastic.
Plastic pollution is increasingly affecting the health of coasts and oceans. One well-known problem is plastic bottles made from polyethylene terephthalate, or PET. A new study involving scientists from Professor Ruth Schmitz-Streit’s research group at Kiel University has shown for the first time, using microorganisms from the deep sea, that polymers such as PET are continuously degraded by an enzyme. Researchers from the University of Hamburg and the Heinrich-Heine-University Düsseldorf played a major role in the microbiological study. The results fundamentally expand the knowledge of PET-degrading enzymes, the underlying mechanism and the evolutionary understanding of the diversity of putative PET-degrading enzymes throughout the global ocean. The research team recently published the results in the journal Communications Chemistry, where they discuss both biotechnological applications and the high relevance for biogeochemical processes in the ocean and on land.
The study highlights a special feature of the PET-degrading enzyme. “In our study, we have discovered a new genetic resource from deep-sea organisms belonging to the archaea,” says Professor Ruth Schmitz-Streit, head of the Molecular Biology of Microorganisms working group at the Institute of General Microbiology (IfAM) and member of the research priority area Kiel Marine Science (KMS) at Kiel University. Until now, about 80 different PET-degrading enzymes were known, most of which were found in bacteria or fungi. “Our data contribute to a better understanding of the ecological role of deep-sea archaea and the possible degradation of PET waste in the sea,” says the microbiologist.
The new enzyme: PET46
Using a metagenomic approach, the research team has identified and biochemically described the PET-degrading enzyme PET46 from a non-cultured deep-sea microorganism for the first time. This involved identifying the gene from a deep-sea sample based on similarities to known sequences, synthesising the corresponding coding gene, producing the protein in the bacterium Escherichia coli and then studying it biochemically and structurally. PET46 has many unusual properties and adds to the scaffold diversity of PET-active enzymes. Structurally, the enzyme differs significantly from those previously discovered. For example, it has the ability to degrade both very long-chain PET molecules, known as polymers, and short-chain PET molecules, known as oligomers, which means that degradation can be continuous.
Among other things, PET46 uses a completely different mechanism for substrate binding than previously known PET-degrading enzymes. The researchers describe an unusual ‘lid’ of 45 amino acids above the enzyme’s active centre as crucial for binding. In other PET enzymes, aromatic amino acids close to the active site are typical.
Promising biotechnology applications
At the molecular level, PET46 is very similar to another enzyme, ferulic acid esterase. This degrades the natural polymer lignin in plant cell walls by breaking down lignin polymers to release sugars from woody plant parts. Lignin and PET have many structural similarities, so the PET-degrading enzymes found in nature may be important for composting wood in forest soils, for example.
The biochemical properties of PET46 therefore make it a very interesting enzyme both for marine and terrestrial plastics and for biotechnology. Compared to the best-characterized PET-degrading enzymes from bacteria and composting plants, PET46 is more efficient at 70 degrees Celsius than these reference enzymes at their respective optimum temperatures.
The research was carried out as part of the PLASTISEA project, coordinated by Professor Ute Hentschel Humeida of the GEOMAR Helmholtz Centre for Ocean Research in Kiel. First author Dr Jennifer Chow from the University of Hamburg and first author Dr Pablo Pérez-Garcia, who works as a research assistant in Schmitz-Streit’s group, contributed equally to the study. The PLASTISEA project is funded by the German Federal Ministry of Education and Research (BMBF) under the activity “New biotechnological processes based on marine resources – BioProMare” 2020-2023 (funding code: 031B0867A).
Original publication:
Perez-Garcia, P., Chow, J., Costanzi, E., (…), Schmitz-Streit, R., and Streit, W. R. An archaeal lid-containing feruloyl esterase degrades polyethylene terephthalate. Commun Chem 6, 193 (2023). DOI: 10.1038/s42004-023-00998-z, https://www.nature.com/articles/s42004-023-00998-z
Photos are available for download:
https://www.uni-kiel.de/de/pressemitteilungen/2023/227-pet46.png
The structure of PET46 is similar to that of known enzymes such as IsPETase and LCC, but has unique features. One example is the unusual ‘lid’, shown in light green.
© Perez-Garcia, P., Chow, J., Costanzi, E. et al. (2023).
https://www.uni-kiel.de/de/pressemitteilungen/2023/227-ruth-schmitz-streit.jpg
Professor Ruth Schmitz-Streit and scientists from her research group at Kiel University were involved in the new study, which described the PET-degrading enzyme PET46 from a non-cultured deep-sea microorganism for the first time.
© Stefan Kolbe
About Kiel Marine Science (KMS)
Kiel Marine Science (KMS), the Center for interdisciplinary marine science at Kiel University, is devoted to excellent and responsible ocean research at the interface between humans and the ocean. The researchers combine their expertise from various natural and social science disciplines to investigate the risks and opportunities that the sea provides for humans. The success of Kiel Marine Science is based on close interdisciplinary cooperation in research and teaching between researchers from seven faculties at Kiel University. Together with actors from outside the scientific community, they work globally and transdisciplinarily on solutions for sustainable use and protection of the ocean.
https://www.uni-kiel.de/en/research/priority-research-areas/kiel-marine-science
More information:
To the working group Molecular Biology of Microorganisms, https://www.mikrobio.uni-kiel.de/de/ag-schmitz-streit/prof-dr-ruth-schmitz-strei…
About Kiel University’s research initiative Ocean Health, https://www.uni-kiel.de/en/research/priority-research-areas/kiel-marine-science/…
About the PLASTISEA project, https://www.geomar.de/en/research/fb3/fb3-ms/projects/plastisea
Scientific Contact:
Prof. Dr. Ruth Schmitz-Streit
Molecular Biology of Microorganisms, Head
Institute of General Microbiology (IfAM)
Kiel University
E-mail: rschmitz@ifam.uni-kiel.de
Telephone: +49/431/880-4334
Press Contact:
Tobias Hahn
Public Outreach / Science Communication
Press, Digital and Science Communication Services and Kiel Marine Science (KMS)
Kiel University
E-mail: thahn@kms.uni-kiel.de
Telephone: +49/431/880-7185
Kiel University
Press, Communication and Marketing, Eva Sittig, Text/editing: Frauke Pescheck, Tobias Hahn
Postal address: D-24098 Kiel, Germany, Telephone: +49 431 880-2104, Fax: +49 431 880-1355
E-mail: presse@uv.uni-kiel.de, Internet: https://www.uni-kiel.de Twitter: https://www.twitter.com/kieluni
Facebook: https://www.facebook.com/kieluni Instagram: https://www.instagram.com/kieluni
Wissenschaftliche Ansprechpartner:
Prof. Dr. Ruth Schmitz-Streit
Molecular Biology of Microorganisms, Head
Institute of General Microbiology (IfAM)
Kiel University
E-mail: rschmitz@ifam.uni-kiel.de
Telephone: +49/431/880-4334
Originalpublikation:
https://www.nature.com/articles/s42004-023-00998-z
Weitere Informationen:
https://www.uni-kiel.de/en/details/news/227-pet46-enzyme-plasticdegradation
Media Contact
All latest news from the category: Life Sciences and Chemistry
Articles and reports from the Life Sciences and chemistry area deal with applied and basic research into modern biology, chemistry and human medicine.
Valuable information can be found on a range of life sciences fields including bacteriology, biochemistry, bionics, bioinformatics, biophysics, biotechnology, genetics, geobotany, human biology, marine biology, microbiology, molecular biology, cellular biology, zoology, bioinorganic chemistry, microchemistry and environmental chemistry.
Newest articles

NASA: Mystery of life’s handedness deepens
The mystery of why life uses molecules with specific orientations has deepened with a NASA-funded discovery that RNA — a key molecule thought to have potentially held the instructions for…

What are the effects of historic lithium mining on water quality?
Study reveals low levels of common contaminants but high levels of other elements in waters associated with an abandoned lithium mine. Lithium ore and mining waste from a historic lithium…

Quantum-inspired design boosts efficiency of heat-to-electricity conversion
Rice engineers take unconventional route to improving thermophotovoltaic systems. Researchers at Rice University have found a new way to improve a key element of thermophotovoltaic (TPV) systems, which convert heat…



