Bulges calculated in the supercomputer

Detail of a membrane bubble.
Image: Bhaskara Group, Goethe University Frankfurt
How cells digest their internal canal system.
Newly discovered mechanism helps detach and recycle parts of cellular canal membranes as needed – models developed using supercomputer simulations.
Inside cells, there exists an extensive system of canals known as the endoplasmic reticulum (ER), which consists of membrane-encased tubes that are partially broken down as needed – for instance in case of a nutrient deficiency. As part of this process, bulges or protrusions form in the membrane, which then pinch off and are recycled by the cell. A study by Goethe University Frankfurt has examined this protrusion process using computer simulations. Its finding: certain structural motifs of proteins in the ER membrane play a central role in this process. The study was carried out as part of the “SCALE – Subcellular Architecture of Life” cluster initiative.
The endoplasmic reticulum functions as a reservoir for calcium and carbohydrates and also serves as the site for the synthesis of various hormones. Cells adjust the expansion and networking of their internal canal system as needed. A process known as ER-phagy (“ER-eating”) plays a central role here. During this process, a part of the membrane of an ER tube bulges out and eventually pinches off into a small vesicle. At the same time, a kind of internal cellular “trash bag”, the autophagosome, forms around it. This then fuses with another container that contains highly reactive enzymes, which “shred” the contents of the “trash bag” and recycle it.
“We have known for several years that specific proteins, known as ER-phagy receptors, play a key role in this process,” explains Dr. Ramachandra Bhaskara from Goethe University’s Institute of Biochemistry II. These receptors are located in the membrane of ER tubes and consist of an anchor that inserts into the membrane. Attached to this anchor are two long protein strands that extend outward from the membrane surface like flexible tentacles. “Using complex simulations in supercomputers, we were recently able to show, together with other research groups, that the anchor causes the membrane to curve,” Bhaskara says, adding that “under certain conditions, this can result in a protrusion. In the current study, we have demonstrated that the filamentous structures increase the likelihood and significantly accelerate the formation of such a bulge.”
Proteins form disordered “tentacles” from amino acids
Most proteins adopt a defined three-dimensional shape after they are produced: some parts form coiled, helical structures, while others fold back and forth like the bellows of an accordion. This gives them a compact, relatively rigid form, which also applies to the anchor region of ER-phagy receptors. The tentacles, however, consist of long chains of amino acids that oscillate back and forth in a largely disordered manner – which is also why they are referred to as “intrinsically disordered (protein) regions” or IDRs for short. These extensive movements require space, which they create by causing the membrane in which they are anchored to bulge. “Added to this is another effect,” emphasizes Dr. Sergio Alejandro Poveda Cuevas, the study’s first author: “The IDRs contain short sequences that can fold back under certain conditions. We were able to show that they do this during the formation of the bulges. They then nestle against the membrane like a scaffold, thereby reinforcing its curvature.”
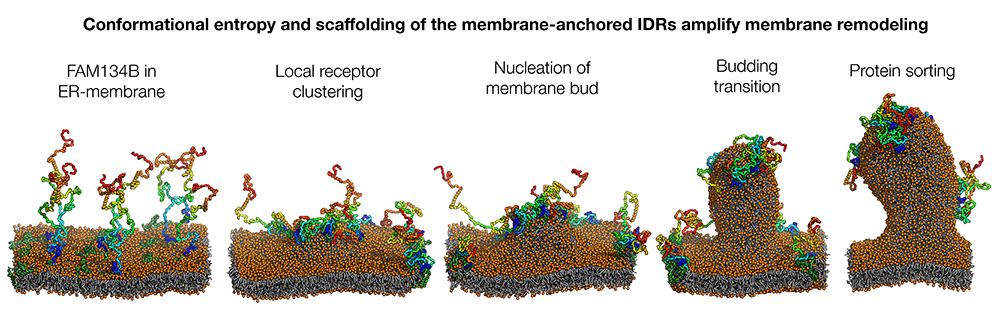
 The ER-phagy receptor FAM134B is initially distributed via the membrane surface (left). Its IDRs move back and forth in a largely disordered manner, like tentacles. The formation of local clusters initiates the curvature of the membrane (2nd from left), which is further strengthened by the condensation of the tentacles into more compact structures (2nd from right). The pinching off of the membrane vesicle (not shown) is initiated by a targeted redistribution of proteins (sorting, right). Image: Bhaskara Group, Goethe University Frankfurt
The ER-phagy receptor FAM134B is initially distributed via the membrane surface (left). Its IDRs move back and forth in a largely disordered manner, like tentacles. The formation of local clusters initiates the curvature of the membrane (2nd from left), which is further strengthened by the condensation of the tentacles into more compact structures (2nd from right). The pinching off of the membrane vesicle (not shown) is initiated by a targeted redistribution of proteins (sorting, right). Image: Bhaskara Group, Goethe University Frankfurt
The pinching-off is thus the result of various finely orchestrated processes, as demonstrated by the simulation: initially, the anchor regions of various ER-phagy receptors approach each other. This clustering increases the curvature of the membrane caused by the receptors. Initially, the IDR tentacles are extended. They make contact with the autophagy machinery and direct it toward the membrane. The IDRs then condense into more compact structures, further enhancing the bulge until the membrane pinches off and the vesicle is packaged in the autophagosome (“trash bag”).
Findings could prove important for the treatment of certain diseases
“In addition to offering a detailed insight into this important cellular process, our study also shows that receptor IDRs play a crucial role in ensuring smooth functioning,” Bhaskara explains. These results are particularly interesting because some congenital neurological diseases are associated with disrupted ER-phagy. A better understanding of the membrane degradation process might one day enable targeted manipulation.
The study was funded by the German Research Foundation (DFG) within the framework of Collaborative Research Center 1177, and by the ENABLE cluster project funded by the Hessian Ministry of Science and Research, Arts and Culture.
Movie/Image for download:
Image sequence: https://www.uni-frankfurt.de/160576923
Movie (19 MB): https://www.uni-frankfurt.de/160576969
Caption (image and movie): The ER-phagy receptor FAM134B is initially distributed via the membrane surface (left). Its IDRs move back and forth in a largely disordered manner, like tentacles. The formation of local clusters initiates the curvature of the membrane (2nd from left), which is further strengthened by the condensation of the tentacles into more compact structures (2nd from right). The pinching off of the membrane vesicle (not shown) is initiated by a targeted redistribution of proteins (sorting, right). Image: Bhaskara Group, Goethe University Frankfurt
Publication:
Sergio Alejandro Poveda-Cuevas, Kateryna Lohachova, Borna Markusic, Ivan Dikic, Gerhard Hummer, Ramachandra M. Bhaskara: Intrinsically disordered region amplifies membrane remodeling to augment selective ER-phagy. PNAS (2024)
Further information
Dr. Ramachandra M. Bhaskara
Head of “Computational Cell Biology” working group
Institute of Biochemistry II
Goethe University Frankfurt
Tel.: +49 (0) 69 6303-2508
bhaskara@med.uni-frankfurt.de
Homepage: https://biochem2.com/research-group/computational-biology
Media Contact
All latest news from the category: Life Sciences and Chemistry
Articles and reports from the Life Sciences and chemistry area deal with applied and basic research into modern biology, chemistry and human medicine.
Valuable information can be found on a range of life sciences fields including bacteriology, biochemistry, bionics, bioinformatics, biophysics, biotechnology, genetics, geobotany, human biology, marine biology, microbiology, molecular biology, cellular biology, zoology, bioinorganic chemistry, microchemistry and environmental chemistry.
Newest articles

NASA: Mystery of life’s handedness deepens
The mystery of why life uses molecules with specific orientations has deepened with a NASA-funded discovery that RNA — a key molecule thought to have potentially held the instructions for…

What are the effects of historic lithium mining on water quality?
Study reveals low levels of common contaminants but high levels of other elements in waters associated with an abandoned lithium mine. Lithium ore and mining waste from a historic lithium…

Quantum-inspired design boosts efficiency of heat-to-electricity conversion
Rice engineers take unconventional route to improving thermophotovoltaic systems. Researchers at Rice University have found a new way to improve a key element of thermophotovoltaic (TPV) systems, which convert heat…
 The ER-phagy receptor FAM134B is initially distributed via the membrane surface (left). Its IDRs move back and forth in a largely disordered manner, like tentacles. The formation of local clusters initiates the curvature of the membrane (2nd from left), which is further strengthened by the condensation of the tentacles into more compact structures (2nd from right). The pinching off of the membrane vesicle (not shown) is initiated by a targeted redistribution of proteins (sorting, right). Image: Bhaskara Group, Goethe University Frankfurt
The ER-phagy receptor FAM134B is initially distributed via the membrane surface (left). Its IDRs move back and forth in a largely disordered manner, like tentacles. The formation of local clusters initiates the curvature of the membrane (2nd from left), which is further strengthened by the condensation of the tentacles into more compact structures (2nd from right). The pinching off of the membrane vesicle (not shown) is initiated by a targeted redistribution of proteins (sorting, right). Image: Bhaskara Group, Goethe University Frankfurt


