Using GPUs to discover human brain connectivity
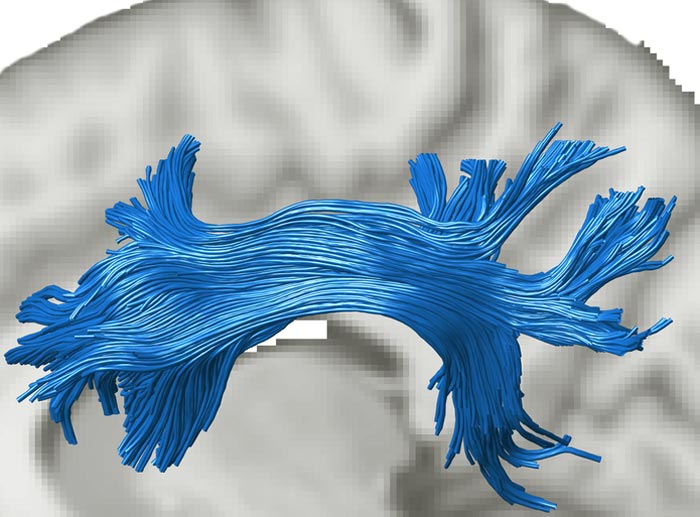
The image shows the superior longitudinal fasciculus (SLF), a white matter tract that connects the prefrontal and parietal cortex, two attention-related brain regions. The tract was estimated with diffusion MRI and tractography in the living human brain
Credit: Varsha Sreenivasan and Devarajan Sridharan
A new GPU-based machine learning algorithm developed by researchers at the Indian Institute of Science (IISc) can help scientists better understand and predict connectivity between different regions of the brain.
The algorithm, called Regularized, Accelerated, Linear Fascicle Evaluation, or ReAl-LiFE, can rapidly analyse the enormous amounts of data generated from diffusion Magnetic Resonance Imaging (dMRI) scans of the human brain. Using ReAL-LiFE, the team was able to evaluate dMRI data over 150 times faster than existing state-of-the-art algorithms.
“Tasks that previously took hours to days can be completed within seconds to minutes,” says Devarajan Sridharan, Associate Professor at the Centre for Neuroscience (CNS), IISc, and corresponding author of the study published in the journal Nature Computational Science.
Millions of neurons fire in the brain every second, generating electrical pulses that travel across neuronal networks from one point in the brain to another through connecting cables or “axons”. These connections are essential for computations that the brain performs. “Understanding brain connectivity is critical for uncovering brain-behaviour relationships at scale,” says Varsha Sreenivasan, PhD student at CNS and first author of the study. However, conventional approaches to study brain connectivity typically use animal models, and are invasive. dMRI scans, on the other hand, provide a non-invasive method to study brain connectivity in humans.
The cables (axons) that connect different areas of the brain are its information highways. Because bundles of axons are shaped like tubes, water molecules move through them, along their length, in a directed manner. dMRI allows scientists to track this movement, in order to create a comprehensive map of the network of fibres across the brain, called a connectome.
Unfortunately, it is not straightforward to pinpoint these connectomes. The data obtained from the scans only provide the net flow of water molecules at each point in the brain. “Imagine that the water molecules are cars. The obtained information is the direction and speed of the vehicles at each point in space and time with no information about the roads. Our task is similar to inferring the networks of roads by observing these traffic patterns,” explains Sridharan.
To identify these networks accurately, conventional algorithms closely match the predicted dMRI signal from the inferred connectome with the observed dMRI signal. Scientists had previously developed an algorithm called LiFE (Linear Fascicle Evaluation) to carry out this optimisation, but one of its challenges was that it worked on traditional Central Processing Units (CPUs), which made the computation time-consuming.
In the new study, Sridharan’s team tweaked their algorithm to cut down the computational effort involved in several ways, including removing redundant connections, thereby improving upon LiFE’s performance significantly. To speed up the algorithm further, the team also redesigned it to work on specialised electronic chips – the kind found in high-end gaming computers – called Graphics Processing Units (GPUs), which helped them analyse data at speeds 100-150 times faster than previous approaches.
This improved algorithm, ReAl-LiFE, was also able to predict how a human test subject would behave or carry out a specific task. In other words, using the connection strengths estimated by the algorithm for each individual, the team was able to explain variations in behavioural and cognitive test scores across a group of 200 participants.
Such analysis can have medical applications too. “Data processing on large scales is becoming increasingly necessary for big-data neuroscience applications, especially for understanding healthy brain function and brain pathology,” says Sreenivasan.
For example, using the obtained connectomes, the team hopes to be able to identify early signs of aging or deterioration of brain function before they manifest behaviourally in Alzheimer’s patients. “In another study, we found that a previous version of ReAL-LiFE could do better than other competing algorithms for distinguishing patients with Alzheimer’s disease from healthy controls,” says Sridharan. He adds that their GPU-based implementation is very general, and can be used to tackle optimisation problems in many other fields as well.
Journal: Nature Computational Science
DOI: 10.1038/s43588-022-00250-z
Article Title: GPU-accelerated connectome discovery at scale
Article Publication Date: 30-May-2022
All latest news from the category: Information Technology
Here you can find a summary of innovations in the fields of information and data processing and up-to-date developments on IT equipment and hardware.
This area covers topics such as IT services, IT architectures, IT management and telecommunications.
Newest articles

Magnetic tornado is stirring up the haze at Jupiter’s poles
Unusual magnetically driven vortices may be generating Earth-size concentrations of hydrocarbon haze. While Jupiter’s Great Red Spot has been a constant feature of the planet for centuries, University of California,…

Cause of common cancer immunotherapy side effect s
New insights into how checkpoint inhibitors affect the immune system could improve cancer treatment. A multinational collaboration co-led by the Garvan Institute of Medical Research has uncovered a potential explanation…

New tool makes quick health, environmental monitoring possible
University of Wisconsin–Madison biochemists have developed a new, efficient method that may give first responders, environmental monitoring groups, or even you, the ability to quickly detect harmful and health-relevant substances…



