AI helping to quantify enzyme activity
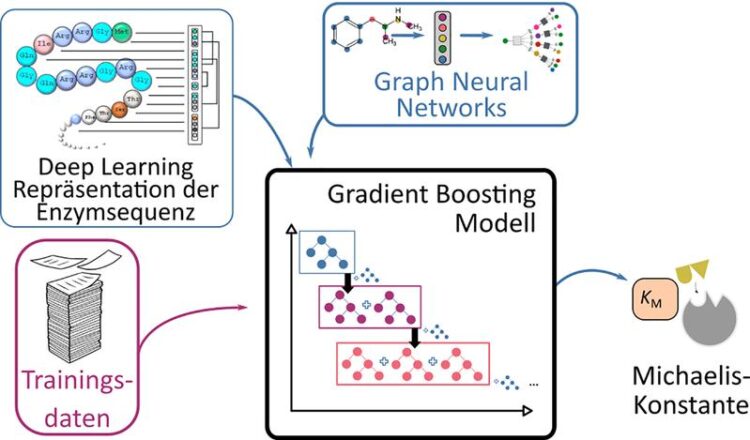
Schematic presentation of the prediction process for Michaelis constants of enzymes using deep learning methods.
Image: HHU / Swastik Mishra
Without enzymes, an organism would not be able to survive. It is these biocatalysts that facilitate a whole range of chemical reactions, producing the building blocks of the cells. Enzymes are also used widely in biotechnology and in our households, where they are used in detergents, for example.
To describe metabolic processes facilitated by enzymes, scientists refer to what is known as the Michaelis-Menten equation. The equation describes the rate of an enzymatic reaction depending on the concentration of the substrate – which is transformed into the end products during the reaction. A central factor in this equation is the ‘Michaelis constant’, which characterises the enzyme’s affinity for its substrate.
It takes a great deal of time and effort to measure this constant in a lab. As a result, experimental estimates of these constants exist for only a minority of enzymes. A team of researchers from the HHU Institute of Computational Cell Biology and Chalmers University of Technology in Stockholm has now chosen a different approach to predict the Michaelis constants from the structures of the substrates and enzymes using AI.
They applied their approach, based on deep learning methods, to 47 model organisms ranging from bacteria to plants and humans. Because this approach requires training data, the researchers used known data from almost 10,000 enzyme-substrate combinations. They tested the results using Michaelis constants that had not been used for the learning process.
Prof. Lercher had this to say about the quality of the results: “Using the independent test data, we were able to demonstrate that the process can predict Michaelis constants with an accuracy similar to the differences between experimental values from different laboratories. It is now possible for computers to estimate a new Michaelis constant in just a few seconds without the need for an experiment.”
The sudden availability of Michaelis constants for all enzymes of model organisms opens up new paths for metabolic computer modelling, as highlighted by the journal PLOS Biology in an accompanying article.
Original publication
Alexander Kroll, Martin K. M. Engqvist, David Heckmann, Martin Lercher, Deep learning allows genome-scale prediction of Michaelis constants from structural features, PLOS Biology (2021).
DOI: 10.1371/journal.pbio.3001402
Journal: PLoS Biology
DOI: 10.1371/journal.pbio.3001402
Article Title: Deep learning allows genome-scale prediction of Michaelis constants from structural features
Article Publication Date: 19-Oct-2021
Media Contact
Arne Claussen
Heinrich-Heine University Duesseldorf
arne.claussen@hhu.de
Office: 211-811-0896
All latest news from the category: Life Sciences and Chemistry
Articles and reports from the Life Sciences and chemistry area deal with applied and basic research into modern biology, chemistry and human medicine.
Valuable information can be found on a range of life sciences fields including bacteriology, biochemistry, bionics, bioinformatics, biophysics, biotechnology, genetics, geobotany, human biology, marine biology, microbiology, molecular biology, cellular biology, zoology, bioinorganic chemistry, microchemistry and environmental chemistry.
Newest articles

You are What You Eat—Stanford Study Links Fiber to Anti-Cancer Gene Modulation
The Fiber Gap: A Growing Concern in American Diets Fiber is well known to be an important part of a healthy diet, yet less than 10% of Americans eat the minimum recommended…

Trust Your Gut—RNA-Protein Discovery for Better Immunity
HIRI researchers uncover control mechanisms of polysaccharide utilization in Bacteroides thetaiotaomicron. Researchers at the Helmholtz Institute for RNA-based Infection Research (HIRI) and the Julius-Maximilians-Universität (JMU) in Würzburg have identified a…

ASXL1 Mutation: The Hidden Trigger Behind Blood Cancers and Inflammation
Scientists show how a mutated gene harms red and white blood cells. LA JOLLA, CA—Scientists at La Jolla Institute for Immunology (LJI) have discovered how a mutated gene kicks off…



