Can proteins bind based only on their shapes?
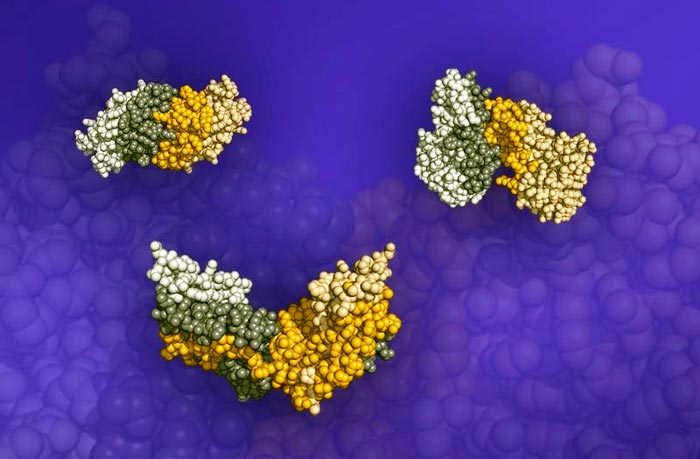
Three protein pair structures. The darkened portions of the structures are the interfaces where the proteins meet and bind.
Image courtesy of Fengyi Gao, University of Michigan and Oak Ridge National Laboratory
A team using the nation’s fastest supercomputer to look at protein binding finds that some binding processes are simpler than expected.
The Science
Proteins bind to one another through a complex mix of chemical interactions. What if some proteins bind due to their shapes, a much simpler process? To answer this question, researchers used Summit, the nation’s fastest supercomputer, to model lock-and-key interactions. In these interactions, molecules in the proteins that are binding together must chemically “fit” precisely. The team tested 46 protein pairs that scientists know bind to one another. Next, the team modeled those protein pairs’ assembly on Summit. Out of the 46 pairs tested, 6 assembled based on their complementary shapes more than 50 percent of the time.
The Impact
The results could have many applications in biological research. For example, the approach could screen drugs for disease treatments or provide scientists with information about how to use proteins as building blocks to design new biological materials. The team plans to study additional proteins that can also bind based on shape—or form even higher order assemblies. Ultimately, the team wants to know the limitation for how protein shapes can evolve to form hierarchical protein structures.
Summary
For proteins to successfully bind to one another, one of them acts as a ligand, a molecule that attaches to a target protein, and one of them acts as a receptor, the molecule that receives the ligand. This process involves complex chemical interactions, in which molecules share bonds and change their configurations upon binding. Researchers wanted to see whether they could predict this binding based on shape alone, ignoring the interactions between proteins. From a database of more than 6,000 protein pairs, the team tested 46 pairs that are known to bind to one another and simulated their assembly on Summit, the Oak Ridge Leadership Computing Facility’s 200-petaflop supercomputer.
The team captured six protein pairs that bound based only on shape, with one of the pairs binding more than 94 percent of the time. The team plans to study more proteins that can also bind based on shape—or form even higher order structures. In the future, the team wants to know the limitation for how protein shapes can evolve to form hierarchical protein structures. The team hopes they can eventually predict the binding of protein-protein interfaces in protein clusters or protein crystallization structures.
Funding
This research used resources of the Oak Ridge Leadership Computing Facility, a Department of Energy Office of Science User Facility. The results described here are based on work supported by the U.S. Army Research Laboratory and the U. S. Army Research Office.
Journal: Soft Matter
DOI. 10.1039/D1SM00468A
Method of Research: Computational simulation/modeling
Article Title: The role of complementary shape in protein dimerization
Article Publication Date: 19-Jul-2021
Media Contact
Michael Church
DOE/US Department of Energy
michael.church@science.doe.gov
Office: 2028416299
All latest news from the category: Life Sciences and Chemistry
Articles and reports from the Life Sciences and chemistry area deal with applied and basic research into modern biology, chemistry and human medicine.
Valuable information can be found on a range of life sciences fields including bacteriology, biochemistry, bionics, bioinformatics, biophysics, biotechnology, genetics, geobotany, human biology, marine biology, microbiology, molecular biology, cellular biology, zoology, bioinorganic chemistry, microchemistry and environmental chemistry.
Newest articles

Parallel Paths: Understanding Malaria Resistance in Chimpanzees and Humans
The closest relatives of humans adapt genetically to habitats and infections Survival of the Fittest: Genetic Adaptations Uncovered in Chimpanzees Görlitz, 10.01.2025. Chimpanzees have genetic adaptations that help them survive…

You are What You Eat—Stanford Study Links Fiber to Anti-Cancer Gene Modulation
The Fiber Gap: A Growing Concern in American Diets Fiber is well known to be an important part of a healthy diet, yet less than 10% of Americans eat the minimum recommended…

Trust Your Gut—RNA-Protein Discovery for Better Immunity
HIRI researchers uncover control mechanisms of polysaccharide utilization in Bacteroides thetaiotaomicron. Researchers at the Helmholtz Institute for RNA-based Infection Research (HIRI) and the Julius-Maximilians-Universität (JMU) in Würzburg have identified a…



