Decoding the structure of the huntingtin protein
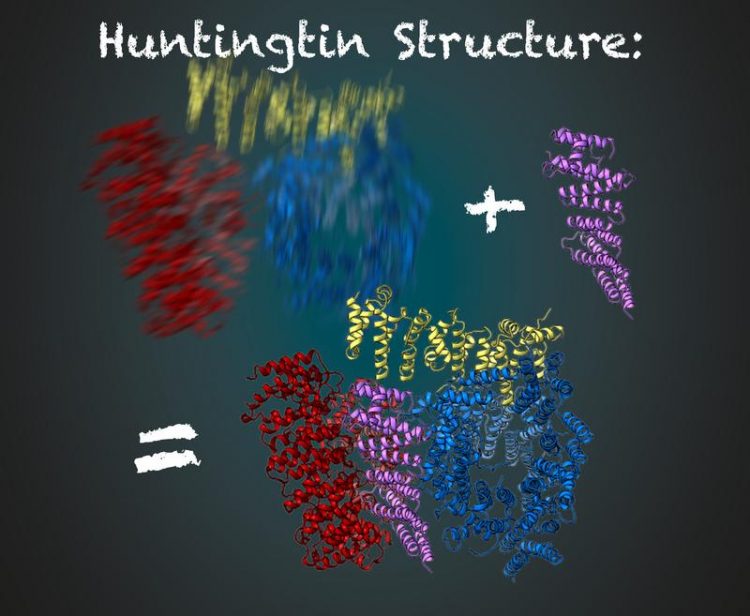
The protein huntingtin consists of three flexible regions (red, yellow and blue). Stabilized by HAP40 (purple), scientists are able to deduce the structure of huntingtin using cryo-electron microscopy Illustration: Gabriele Stautner, ARTIFOX © Max Planck Institute of Biochemistry
Huntington's disease often begins with emotional disturbances and is characterized by involuntary muscle movement and loss of mental ability. Until today, the neurological disease is one of the hereditary diseases still incurable and fatal. The protein HTT, also called huntingtin, plays the central role in Huntington's disease. For 25 years it has been known that mutations of the huntingtin gene, which is the blueprint for the same protein, cause Huntington's disease.
Although researchers have worked on it for many years, there are still many hurdles to overcome. Now Rubén Fernández-Busnadiego from the Department of Molecular Structural Biology at the MPI of Biochemistry and Stefan Kochanek, head of the Department of Gene Therapy at the University Hospital Ulm, have succeeded in decoding the molecular, three-dimensional structure of the huntingtin protein.
Overcome hurdle
Stefan Kochanek and his team have been working on the production and purification of huntingtin for a long time. What has prevented a detailed analysis of the protein in recent decades? Fernández-Busnadiego, an expert in cryo-electron microscopy, mentions two main factors: “First of all, cryo-electron microscopy has only been optimized in recent years to look at protein structures with almost molecular resolution. The second reason is that the huntingtin protein is very flexible in its structure. Just now, we have found also a solution for this problem.”
During the analysis, pictures of the protein are being taken from different perspectives under the microscope. The three-dimensional molecular structure can be computed from the large number of resulting images. For this, the protein must always be in the same conformation. Fernández-Busnadiego explains: “This would be similar to a person being photographed in the dark. If the person does not stand still for a while, the shot will be blurry.”
To get a clear picture, the researchers in the Kochanek lab have been looking for other proteins that interact with huntingtin and stabilize it. That worked for the protein HAP40. “Huntingtin in connection with HAP40 is stabilized in a particular conformation. Thus, averaged over many pictures, we were able to derive the three-dimensional structure,” said Kochanek. “If we remain in the analogy of the photo in the dark, then the protein acts like a chair for the photographed person. Sitting on it, the person moves much less and the image is much sharper with the same exposure time,” adds Fernández-Busnadiego.
Why is the three-dimensional structure of huntingtin needed?
“Although we have known for some time that the mutation of the huntingtin gene has severe consequences, we still know relatively little about the function and tasks of the healthy protein,” explains Kochanek. Proteins are the molecular machines of the cell. In order to fulfill their versatile tasks, they have a certain three-dimensional structure, similar to a specific component in a machine.
“Now that we know the exact structure of huntingtin, we can further study which areas of huntingtin are particularly important and how other proteins cooperate with huntingtin functionally. In this way structures could be deduced to be targeted therapeutically by certain drugs.”
Right now, there is a lot going on in Huntington's disease research. Great hope is directed to a method, that mutes the huntingtin gene with so-called Antisense-Oligonukleotiden (ASO) for the treatment of Huntington's disease. These small molecules reduce the formation of huntingtin proteins in the cells, but the drug cannot differentiate between the normal and the pathologically altered huntingtin protein. Also for this reason, it is important to learn more about the function of the healthy huntingtin protein. Kochanek confidently looks into the future: “The decoded structure will bring us a big step ahead”.
[CM]
Comment from the clinician
Bernhard Landwehrmeyer is the Director of the Huntington Outpatient Clinic of the Neurology Department at the University Medical Center Ulm and heads the world's largest international cohort study on Huntington's disease (with more than 16,000 participants): “Identifying the structure of huntingtin is a huge step forward for the many families hoping for effective treatment for Huntington's disease. The illness partly arises due to new characteristics of the pathologically changed huntingtin gene products that are the result of the HD mutation, but also in part as a consequence of impaired normal functioning of huntingtin. Now – armed with the knowledge of the architecture of huntingtin – questions about the normal function of huntingtin can be clarified in a much quicker and more tightly focused way, and tailored drugs can be developed that promote normal function.”
“The antisense oligonucleotide (ASO) treatment trials are presently only conducted at very few clinics; the Neurological University Clinic Ulm leads the drug testing for Germany. Some of the currently reviewed ASOs reduce both the formation of the normal and the mutant huntingtin protein, while other ASOs predominantly seek to lower the altered huntingtin. It is currently unclear whether a partial inhibition of the formation of normal huntingtin protein has disadvantages and is tolerated without undesirable side effects. This is yet another reason why it is important to learn more about the normal function of the huntingtin protein. To this end, the study published in the journal Nature will make an important contribution.”
Original publication:
Q. Guo, B. Huang, J. Cheng, M. Seefelder, T. Engler, G. Pfeifer, P. Oeckl, M. Otto, F. Moser, M. Maurer, A. Pautsch, W. Baumeister, R Fernández-Busnadiego and S. Kochanek: The cryo-EM structure of huntingtin, Nature, February 2018
DOI: 10.1038 / nature25502
Contact:
Dr. Rubén Fernández-Busnadiego
Molecular Structural Biology
Max-Planck-Institut für Biochemie
Am Klopferspitz 18
82152 Martinsried
E-Mail: ruben@biochem.mpg.de
http://www.biochem.mpg.de/en/rd/baumeister
Dr. Christiane Menzfeld
Public Relations
Max-Planck-Institut für Biochemie
Am Klopferspitz 18
82152 Martinsried
Tel. +49 89 8578-2824
E-Mail: pr@biochem.mpg.de
http://www.biochem.mpg.de/en
Prof. Dr. Stefan Kochanek
Abteilung Gentherapie
Universitätsklinikum Ulm
Helmholtzstr. 8/1
89081 Ulm
E-Mail: stefan.kochanek@uni-ulm.de
https://www.uniklinik-ulm.de/abteilung-gentherapie.html
Annika Bingmann
Presse- und Öffentlichkeitsarbeit
Universität Ulm
Helmholtzstraße 16
89081 Ulm
Tel.: 0731-50 22121
Fax.: 0731-50 22048
E-Mail: annika.bingmann@uni-ulm.de
http://www.uni-ulm.de/universitaet/hochschulkommunikation/presse-und-oeffentlich…
http://www.biochem.mpg.de/en/news/pressroom – More press releases of the MPI of Biochemistry
http://www.biochem.mpg.de/en/rd/baumeister – Website of the research department „”Molecular Structural Biology”
https://twitter.com/MPI_Biochem – Twitter @MPI_Biochem
Media Contact
All latest news from the category: Life Sciences and Chemistry
Articles and reports from the Life Sciences and chemistry area deal with applied and basic research into modern biology, chemistry and human medicine.
Valuable information can be found on a range of life sciences fields including bacteriology, biochemistry, bionics, bioinformatics, biophysics, biotechnology, genetics, geobotany, human biology, marine biology, microbiology, molecular biology, cellular biology, zoology, bioinorganic chemistry, microchemistry and environmental chemistry.
Newest articles

Hyperspectral imaging lidar system achieves remote plastic identification
New technology could remotely identify various types of plastics, offering a valuable tool for future monitoring and analysis of oceanic plastic pollution. Researchers have developed a new hyperspectral Raman imaging…

SwRI awarded $26 million to develop NOAA magnetometers
SW-MAG data will help NOAA predict, mitigate the effects of space weather. NASA and the National Oceanic and Atmospheric Administration (NOAA) recently awarded Southwest Research Institute a $26 million contract…

Protein that helps cancer cells dodge CAR T cell therapy
Discovery could lead to new treatments for blood cancer patients currently facing limited options. Scientists at City of Hope®, one of the largest and most advanced cancer research and treatment…



