Deep learning techniques teach neural model to 'play' retrosynthesis
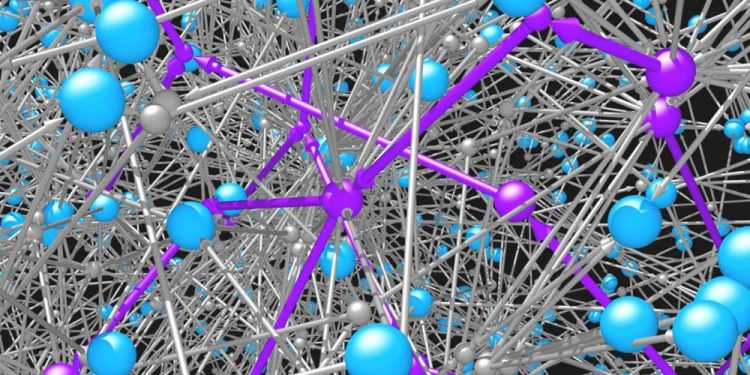
Molecules (blue spheres) are connected to one another by the reactions (grey spheres and arrows) in which they participate. The network of possible organic molecules and reactions is impossibly vast. Intelligent search algorithms are needed to identify feasible pathways (purple) for synthesizing desired molecules. Credit: Mikolaj Kowalik & Kyle Bishop/Columbia Engineering
Researchers, from biochemists to material scientists, have long relied on the rich variety of organic molecules to solve pressing challenges. Some molecules may be useful in treating diseases, others for lighting our digital displays, still others for pigments, paints, and plastics.
The unique properties of each molecule are determined by its structure–that is, by the connectivity of its constituent atoms. Once a promising structure is identified, there remains the difficult task of making the targeted molecule through a sequence of chemical reactions. But which ones?
Organic chemists generally work backwards from the target molecule to the starting materials using a process called retrosynthetic analysis. During this process, the chemist faces a series of complex and inter-related decisions. For instance, of the tens of thousands of different chemical reactions, which one should you choose to create the target molecule?
Once that decision is made, you may find yourself with multiple reactant molecules needed for the reaction. If these molecules are not available to purchase, then how do you select the appropriate reactions to produce them? Intelligently choosing what to do at each step of this process is critical in navigating the huge number of possible paths.
Researchers at Columbia Engineering have developed a new technique based on reinforcement learning that trains a neural network model to correctly select the “best” reaction at each step of the retrosynthetic process.
This form of AI provides a framework for researchers to design chemical syntheses that optimize user specified objectives such synthesis cost, safety, and sustainability. The new approach, published May 31 by ACS Central Science, is more successful (by ~60%) than existing strategies for solving this challenging search problem.
“Reinforcement learning has created computer players that are much better than humans at playing complex video games. Perhaps retrosynthesis is no different! This study gives us hope that reinforcement-learning algorithms will be perhaps one day better than human players at the 'game' of retrosynthesis,” says Alán Aspuru-Guzik, professor of chemistry and computer science at the University of Toronto, who was not involved with the study.
The team framed the challenge of retrosynthetic planning as a game like chess and Go, where the combinatorial number of possible choices is astronomical and the value of each choice uncertain until the synthesis plan is completed and its cost evaluated. Unlike earlier studies that used heuristic scoring functions–simple rules of thumb–to guide retrosynthetic planning, this new study used reinforcement learning techniques to make judgments based on the neural model's own experience.
“We're the first to apply reinforcement learning to the problem of retrosynthetic analysis,” says Kyle Bishop, associate professor of chemical engineering. “Starting from a state of complete ignorance, where the model knows absolutely nothing about strategy and applies reactions randomly, the model can practice and practice until it finds a strategy that outperforms a human-defined heuristic.”
In their study, Bishop's team focused on using the number of reaction steps as the measurement of what makes a “good” synthetic pathway. They had their reinforcement learning model tailor its strategy with this goal in mind. Using simulated experience, the team trained the model's neural network to estimate the expected synthesis cost or value of any given molecule based on a representation of its molecular structure.
The team plans to explore different goals in the future, for instance, training the model to minimize costs rather than the number of reactions, or to avoid molecules that could be toxic. The researchers are also trying to reduce the number of simulations required for the model to learn its strategy, as the training process was quite computationally expensive.
“We expect that our retrosynthesis game will soon follow the way of chess and Go, in which self-taught algorithms consistently outperform human experts,” Bishop notes. “And we welcome competition. As with chess-playing computer programs, competition is the engine for improvements in the state-of-the-art, and we hope that others can build on our work to demonstrate even better performance.”
###
About the Study
The study is titled “Learning retrosynthetic planning through simulated experience.”
Authors are: John S. Schreck and Kyle J. M. Bishop, Chemical Engineering, Columbia Engineering, Connor W. Coley, Chemical Engineering, Massachusetts Institute of Technology.
The study was supported by the DARPA Make-It program under contract ARO W911NF-16- 2-0023.
The authors declare no financial or other conflicts of interest.
LINKS:
Paper: https:/
DOI: http://dx.
https:/
http://engineering.
https:/
https:/
Columbia Engineering
Columbia Engineering, based in New York City, is one of the top engineering schools in the U.S. and one of the oldest in the nation. Also known as The Fu Foundation School of Engineering and Applied Science, the School expands knowledge and advances technology through the pioneering research of its more than 220 faculty, while educating undergraduate and graduate students in a collaborative environment to become leaders informed by a firm foundation in engineering. The School's faculty are at the center of the University's cross-disciplinary research, contributing to the Data Science Institute, Earth Institute, Zuckerman Mind Brain Behavior Institute, Precision Medicine Initiative, and the Columbia Nano Initiative. Guided by its strategic vision, “Columbia Engineering for Humanity,” the School aims to translate ideas into innovations that foster a sustainable, healthy, secure, connected, and creative humanity.
Media Contact
More Information:
http://dx.doi.org/10.1021/acscentsci.9b00055All latest news from the category: Life Sciences and Chemistry
Articles and reports from the Life Sciences and chemistry area deal with applied and basic research into modern biology, chemistry and human medicine.
Valuable information can be found on a range of life sciences fields including bacteriology, biochemistry, bionics, bioinformatics, biophysics, biotechnology, genetics, geobotany, human biology, marine biology, microbiology, molecular biology, cellular biology, zoology, bioinorganic chemistry, microchemistry and environmental chemistry.
Newest articles

Hyperspectral imaging lidar system achieves remote plastic identification
New technology could remotely identify various types of plastics, offering a valuable tool for future monitoring and analysis of oceanic plastic pollution. Researchers have developed a new hyperspectral Raman imaging…

SwRI awarded $26 million to develop NOAA magnetometers
SW-MAG data will help NOAA predict, mitigate the effects of space weather. NASA and the National Oceanic and Atmospheric Administration (NOAA) recently awarded Southwest Research Institute a $26 million contract…

Protein that helps cancer cells dodge CAR T cell therapy
Discovery could lead to new treatments for blood cancer patients currently facing limited options. Scientists at City of Hope®, one of the largest and most advanced cancer research and treatment…



