Evolutionary model predicts partitioning of molecules within cells
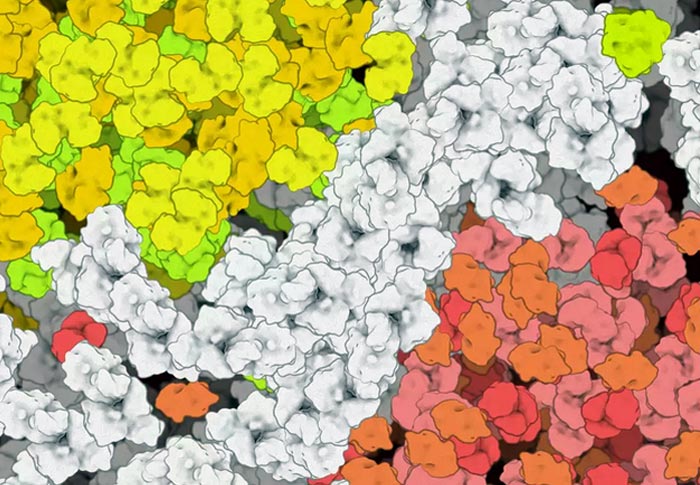
Schematic illustration of densely packed molecules in a cell. Interactions between many different molecules result in two types of droplets (yellow and red), embedded in the background fluid (white). Picture created using cellPAINT.
© MPIDS, Zwicker
Researchers from the Max Planck Institute for Dynamics and Self-Organization (MPI-DS) in Göttingen, Germany, and Delft University of Technology, the Netherlands, have developed a new theoretical method to study mixtures consisting of many different molecules. They analyzed how the molecules interact to reliably form different droplets, as happens continuously in living cells. With this model, particular droplet formation based on many interacting molecules can be predicted for the first time. The findings were recently published in the scientific journal PNAS.
Theoretical physicist David Zwicker and biophysicist Liedewij Laan are interested in figuring out the mysteries of life, specifically the fundamental workings of living cells. “I’m always surprised that cellular processes work at all, because a cell consists of thousands of different molecules that all need to work together without interfering with one another,” Zwicker says. The scientists developed a computer model that allows to investigate complex fluids, like the liquid inside of a cell. “Despite their complex chemical and physical nature, molecules within cells are often organized as droplets,” Laan adds. “With our approach, we are able to reproduce the formation of these droplets, and thus opened up new lines of investigation on how this mechanism works”, she continues.
A complex vinaigrette
Biological cells are incredibly complex machines that rely on thousands of different molecules to work and interact reliably. To orchestrate the molecules, they are separated into different compartments within cells. Since the environment and the internal state of cells constantly changes, they have to be formed and maintained robustly. Most well-known compartments, including the nucleus and mitochondria, are enclosed by membranes that serve as barriers defining their shape and controlling their composition. However, many smaller compartments do not possess a membrane and thus often behave more dynamically. Such compartments have been found in animal, plant, and bacterial cells alike. They can form spontaneously and the interactions between the involved molecules control their composition.
To understand how a plethora of different molecules organizes in such membrane-less compartments, Zwicker and Laan theoretically studied the physical process of droplet formation. So far, the underlying theory was only well-understood for the simple case of two particle types, such as oil separating from the water in vinaigrette. To examine a more realistic cellular environment with a larger number of molecular interactions, the researchers introduced a numerical method, which predicts droplet compositions.
Secrets of life
The new numerical approach opens the possibility to answer key questions, including how many different droplets are formed in a robust manner. “The key insight is that during billions of years of evolution the interactions between cellular molecules have been optimized to form the right droplets,” Zwicker explains. To mimic this in the computer, the researchers adjusted the interactions using multiple generations of mutation and selection until a certain number of droplets form. Interestingly, many different sets of interactions almost perfectly end up at the pre-set number of different droplets. Laan comments: “We were excited to find that we can actually reproduce these mixtures. Yet, scientifically even more exciting is perhaps the fact that we do not understand how this works exactly. But now we are now a lot closer to trying to figure out these secrets of life.”
Zwicker emphasizes that their research is part of a bigger paradigm shift in biology: “So far, researchers tended to investigate rather strong interactions, for example between two proteins, as these are quite robust and thus easier to study. Nowadays, droplets that are formed due to much weaker interactions are being studied more and more, both theoretically and experimentally, because them seem to fulfil an important role inside cells. However, they are much more challenging to analyze, as these weak interactions can be perturbed by temperature, acidity, salt concentration, and many other factors.” The present work suggests that complex behavior, like droplet formation, can robustly emerge from even such weak interactions.
This press release was written in collaboration with TU Delft
Originalpublikation:
https://www.pnas.org/doi/abs/10.1073/pnas.2201250119
Weitere Informationen:
Media Contact
All latest news from the category: Life Sciences and Chemistry
Articles and reports from the Life Sciences and chemistry area deal with applied and basic research into modern biology, chemistry and human medicine.
Valuable information can be found on a range of life sciences fields including bacteriology, biochemistry, bionics, bioinformatics, biophysics, biotechnology, genetics, geobotany, human biology, marine biology, microbiology, molecular biology, cellular biology, zoology, bioinorganic chemistry, microchemistry and environmental chemistry.
Newest articles

Innovative 3D printed scaffolds offer new hope for bone healing
Researchers at the Institute for Bioengineering of Catalonia have developed novel 3D printed PLA-CaP scaffolds that promote blood vessel formation, ensuring better healing and regeneration of bone tissue. Bone is…

The surprising role of gut infection in Alzheimer’s disease
ASU- and Banner Alzheimer’s Institute-led study implicates link between a common virus and the disease, which travels from the gut to the brain and may be a target for antiviral…

Molecular gardening: New enzymes discovered for protein modification pruning
How deubiquitinases USP53 and USP54 cleave long polyubiquitin chains and how the former is linked to liver disease in children. Deubiquitinases (DUBs) are enzymes used by cells to trim protein…



