Floppy but fast
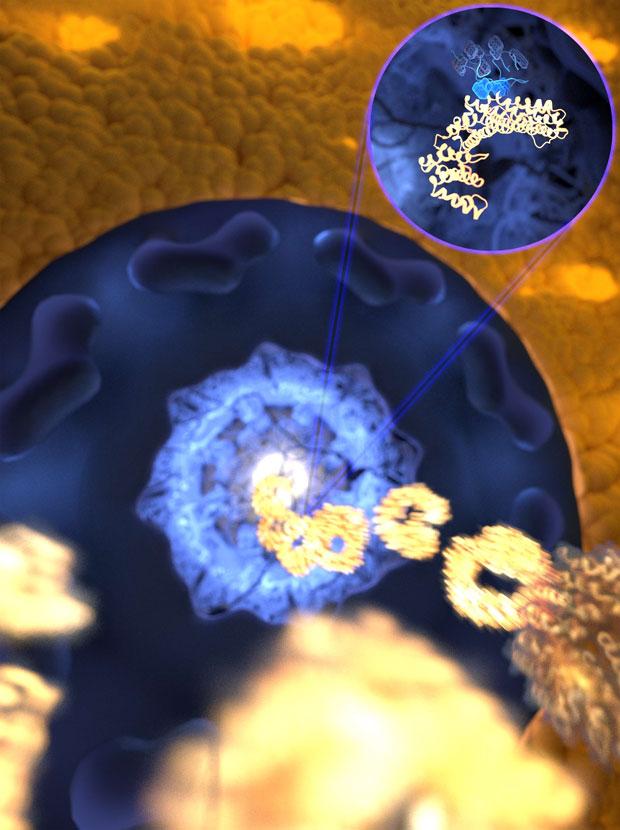
The ultrafast and yet selective binding allows the receptor (gold) to rapidly travel through the pore filled with disordered proteins (blue) into the nucleus, while any unwanted molecules are kept outside. Credit: Mercadante /HITS
Through a combination of computer simulations and various experimental techniques, researchers from Germany, France and the UK have solved this puzzle. A very flexible and disordered protein can bind to its receptor within billionths of a second. Their research, led by Edward Lemke at EMBL, Frauke Gräter at the Heidelberg Institute for Theoretical Studies, and Martin Blackledge at Institut de Biologie Structurale, is published in Cell this week.
Proteins can recognize one another. Each engages very specifically with only a subset of the many different proteins present in the living cell, like a key slotting into a lock. But what if the key is completely flexible, as is the case for so-called intrinsically disordered proteins (IDPs)?
The research teams headed by Edward Lemke at EMBL Heidelberg, Frauke Gräter at the Heidelberg Institute for Theoretical Studies (HITS) and Martin Blackledge at the Institut de Biologie Structurale (IBS) in France, addressed this question in a highly interdisciplinary collaboration, combining molecular simulations, single molecule fluorescence resonance energy transfer (FRET), nuclear magnetic resonance (NMR), stopped flow spectroscopy and in-cell particle tracking.
Unexpectedly, they found that flexible, spaghetti-like proteins can be good – maybe even better than solid protein blocks – at being recognised by multiple partners. And they can do so very fast, while still retaining the high specificity the cell needs. In fact, this could be why these disordered molecules are more common in evolutionarily higher organisms, the researchers surmise.
Researchers had assumed that when an IDP 'key' needed to bind to its lock, it rearranged itself to become more rigid, but experiments in the Lemke lab hinted otherwise. “The pioneering single molecule experiments undertaken at EMBL showed for the particular interaction of a receptor with a disordered protein no hint of rigidity: the flexible protein stayed as flexible even when bound to its receptor” says Davide Mercadante (HITS).
This prompted him to study the very same interaction on the computer. The surprising result was that the high flexibility of the IDP actually helps it bind to its lock – in this case, a nuclear transport receptor, which shuttles proteins into the nucleus. The simulations even suggested the binding to be ultrafast – faster than any other association of that kind recorded to date.
“The computational data indicated that we might have identified a new ultrafast binding mechanism, but it took us three years to design experiments to prove the kinetics in the lab,” Iker Valle Aramburu (EMBL) recalls. “In the end, we had a remarkably perfect match.”
The results now help to understand a long-standing paradox: “For a cell to be viable, molecules must constantly move into and out of its nucleus”, says Edward Lemke (EMBL). “Our findings explain the so-called transport paradox – that is, how this shuttling can be so very fast while remaining specific so that unwanted molecules cannot pass the barrier that protects our genome.”
The new study suggests that many binding motifs at the surface of the IDP create a highly reactive surface that together with the very high speed of locking and unlocking ensures efficient proof-reading while the receptors to travel so fast through a pore filled with other IDPs.
“This is likely a new paradigm for the recognition of intrinsically disordered proteins.” says Frauke Gräter (HITS). Since around 30-50% of the proteins in human cells are disordered, at least in some regions of the protein, the results may also provide a rationale for how recognition information can be processed very fast in general – which is vital to cells.
Other researchers involved in the study are working at the IBS in Grenoble, France, and Cambridge University, UK.
Media Contact
All latest news from the category: Life Sciences and Chemistry
Articles and reports from the Life Sciences and chemistry area deal with applied and basic research into modern biology, chemistry and human medicine.
Valuable information can be found on a range of life sciences fields including bacteriology, biochemistry, bionics, bioinformatics, biophysics, biotechnology, genetics, geobotany, human biology, marine biology, microbiology, molecular biology, cellular biology, zoology, bioinorganic chemistry, microchemistry and environmental chemistry.
Newest articles

NASA: Mystery of life’s handedness deepens
The mystery of why life uses molecules with specific orientations has deepened with a NASA-funded discovery that RNA — a key molecule thought to have potentially held the instructions for…

What are the effects of historic lithium mining on water quality?
Study reveals low levels of common contaminants but high levels of other elements in waters associated with an abandoned lithium mine. Lithium ore and mining waste from a historic lithium…

Quantum-inspired design boosts efficiency of heat-to-electricity conversion
Rice engineers take unconventional route to improving thermophotovoltaic systems. Researchers at Rice University have found a new way to improve a key element of thermophotovoltaic (TPV) systems, which convert heat…



