Mapping E. coli to overcome antibiotic resistance
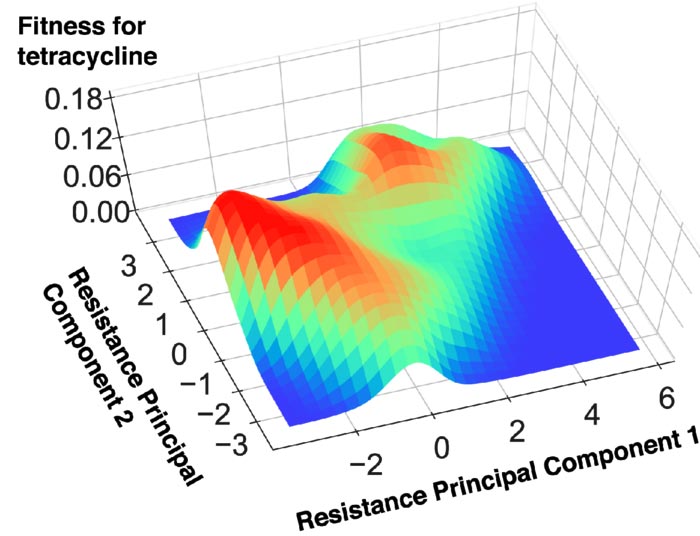
This topographical “map” shows the step-by-step development of E. coli’s resistance to tetracycline, a common antibiotic used to treat a wide variety of infections. The bacteria at the peak of the “mountains” (in red) have the best fitness and most likely chance of resisting the effects of the antibiotic.
Credit: Modified from 2022 Iwasawa et al.
Charts called fitness landscapes could help researchers better understand the evolution of antibiotic-resistant bacteria and find treatments.
Antibiotic resistance, when infection-causing bacteria evolve so they are no longer affected by typical antibiotics, is a global concern. New research at the University of Tokyo has mapped the evolution and process of natural selection of Escherichia coli (E. coli) bacteria in the lab. These maps, called fitness landscapes, help us better understand the step-by-step development and characteristics of E. coli resistance to eight different drugs, including antibiotics. Researchers hope their results and methods will be useful for predicting and controlling E. coli and other bacteria in the future.
Have you ever felt queasy after eating an undercooked burger? Or when leftovers from yesterday’s dinner were left out of the fridge a bit too long? There are many different kinds of food poisoning, but one common cause is the growth of bacteria such as E. coli. Most cases of E. coli, though unpleasant, can be managed at home with rest and rehydration. However, in some instances, it can lead to life-threatening infections. If you have a bacterial infection, antibiotic medication can be a powerful and effective treatment. But antibiotic resistance, the ability of bacteria to become strong enough that it does not respond to the medication, is a serious global concern. If antibiotics are no longer effective, then we will once again be at risk of serious illness from small injuries and common ailments.
“The development of methods that could predict and control bacterial evolution is crucial to find and suppress the emergence of resistant bacteria,” said researcher Junichiro Iwasawa, a doctoral student in the Graduate School of Science at the time of the study. “Thus, we have developed a novel method to predict drug resistance evolution by using data obtained from laboratory evolution experiments of E. coli.”
The researchers used a method called adaptive laboratory evolution, or ALE, to “replay the tape” on the evolution of drug-resistant E. coli to eight different drugs, including antibiotics. The method enabled the researchers to study the evolution of bacterial strains with specific observable characteristics (called phenotypes) in the lab. This helped them gain insight into what changes might occur to the bacteria during the longer-term process of natural selection.
“While conventional laboratory evolution experiments have been labor intensive, we mitigated this problem by using an automated culture system that was previously developed in our lab. This allowed us to acquire sufficient data on the phenotypic changes related to drug resistance evolution,” explained Iwasawa. “By analyzing the acquired data, using principal component analysis (a machine-learning method), we have been able to elucidate the fitness landscape which underlies the drug resistance evolution of E. coli.”
Fitness landscapes look like 3D topographic maps. The mountains and valleys on the map represent an organism’s evolution. Organisms at the peaks have evolved to have better “fitness,” or ability to survive in their environment. Iwasawa explained, “The coordinates of the fitness landscape represent inner states of the organism, such as gene mutation patterns (genotypes) or drug resistance profiles (phenotypes), etc. Thus, the fitness landscape describes the relation between the inner states of the organism and its corresponding fitness levels. By elucidating the fitness landscape, the progression of evolution is expected to be predictable.”
The team believes the fitness landscapes it has mapped in this study and the methods developed in the process will be useful for predicting and controlling not only E. coli, but also other forms of microbial evolution. The researchers hope this will lead to future studies that can find ways to suppress drug-resistant bacteria and contribute to the development of useful microbes for bioengineering and agriculture. Iwasawa concluded that “the next important step is to actually try using the fitness landscapes to control drug resistance evolution and see how far we can control it. This can be done by designing laboratory evolution experiments based on the information from the landscapes. We can’t wait to see the upcoming results.”
Paper Title:
Junichiro Iwasawa, Tomoya Maeda, Atsushi Shibai, Hazuki Kotani, Masako Kawada, Chikara Furusawa “Analysis of the evolution of resistance to multiple antibiotics enables prediction of the Escherichia coli phenotype-based fitness landscape”. PLoS Biology. 2022; 20(12): e3001920. https://doi.org/10.1371/journal.pbio.3001920.
Funding:
This work was supported in part by JSPS KAKENHI (17H06389 and 19H05626 to C.F.) and JST ERATO (JPMJER1902 to C.F).
Useful Links:
Graduate School of Science: https://www.s.u-tokyo.ac.jp/en/info/6799/
RIKEN Center for Biosystems Dynamics Research: https://www.bdr.riken.jp/en/index.html
Furusawa Lab: http://www.furusawa-lab.ubi.s.u-tokyo.ac.jp/en/
Research contact:
Professor Chikara Furusawa
Universal Biology Institute,
Graduate School of Science, The University of Tokyo,
7-3-1 Hongo, Bunkyo-ku, Tokyo 113-0033, Japan
Email: furusawa@phys.s.u-tokyo.ac.jp
Press contact:
Mrs. Nicola Burghall
Public Relations Group, The University of Tokyo,
7-3-1 Hongo, Bunkyo-ku, Tokyo 113-8654, Japan
press-releases.adm@gs.mail.u-tokyo.ac.jp
About the University of Tokyo
The University of Tokyo is Japan’s leading university and one of the world’s top research universities. The vast research output of some 6,000 researchers is published in the world’s top journals across the arts and sciences. Our vibrant student body of around 15,000 undergraduate and 15,000 graduate students includes over 4,000 international students. Find out more at www.u-tokyo.ac.jp/en/ or follow us on Twitter at @UTokyo_News_en.
Journal: PLoS Biology
DOI: 10.1371/journal.pbio.3001920
Method of Research: Experimental study
Subject of Research: Cells
Article Title: Analysis of the evolution of resistance to multiple antibiotics enables prediction of the Escherichia coli phenotype-based fitness landscape
Article Publication Date: 13-Dec-2022
Original Source
All latest news from the category: Life Sciences and Chemistry
Articles and reports from the Life Sciences and chemistry area deal with applied and basic research into modern biology, chemistry and human medicine.
Valuable information can be found on a range of life sciences fields including bacteriology, biochemistry, bionics, bioinformatics, biophysics, biotechnology, genetics, geobotany, human biology, marine biology, microbiology, molecular biology, cellular biology, zoology, bioinorganic chemistry, microchemistry and environmental chemistry.
Newest articles

A new class of cosmic X-ray sources discovered
An international team of astronomers, led by researchers from the Astronomical Observatory of the University of Warsaw, have identified a new class of cosmic X-ray sources. The findings have been…

An open solution to improving research reproducibility
Academic and industry scientists collaborate on a new method to characterize research antibodies. Structural Genomics Consortium researchers at The Neuro (Montreal Neurological Institute-Hospital) of McGill University, in collaboration with scientists…

Living in the deep, dark, slow lane
Insights from the first global appraisal of microbiomes in earth’s subsurface environments. Which microbes thrive below us in darkness – in gold mines, in aquifers, in deep boreholes in the…



