Mapping of Genetic Control Elements in the Cerebellum
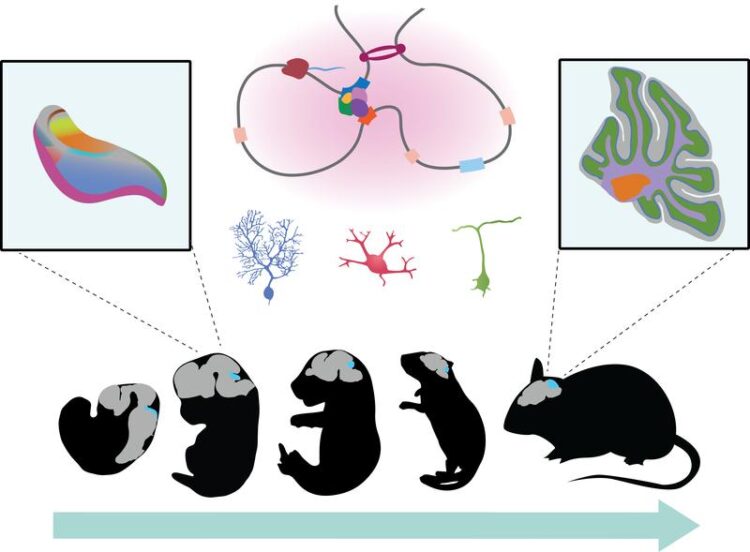
Atlas of genetic control elements in the cerebellum: most control elements are specific for individual cell types and development stages.
(c) Research group Prof. Dr Henrik Kaessmann
The mammalian cerebellum has long been associated almost exclusively with motor control, yet recent studies indicate that it also contributes to many higher brain functions. An international research team led by Prof. Dr Henrik Kaessmann from the Center for Molecular Biology of Heidelberg University (ZMBH) has now decoded the genetic programmes that control the development of cerebellar cell types before and after birth.
The molecular biologists compared data from the mouse cerebellum with corresponding data from the opossum, revealing fundamental gene regulatory networks that must have already formed in the early stage of mammalian evolution more than 160 million years ago. The study was carried out in close collaboration with Prof. Dr Stefan Pfister of the Hopp Children’s Cancer Center Heidelberg (KiTZ).
The development of mammalian organs is controlled by the finely tuned activation and complex interaction of many different genes – also known as gene expression networks. “Although developmental gene expression in the cerebellum has already been studied relatively well, its genetic control has remained elusive,” explains Dr Mari Sepp, a postdoctoral researcher in Prof. Kaessmann’s “Functional evolution of mammalian genomes” research group. In the current study the researchers mapped, at the cellular level, the control elements of all active genes over the entire developmental period of the cerebellum in mice. They used state-of-the-art single-cell sequencing techniques for this task.
On the basis of these data and bioinformatic analysis methods, they were then able to decode the regulatory programmes that control the gene expression networks of all cells and hence the development of the cerebellum. The researchers identified more than 200,000 control elements, most of which are highly specific for individual cell types and development stages. However, some of these elements are activated in multiple cell types, especially in early stages of development.
To also understand the evolution of these gene regulatory programmes, the researchers compared their results from the mouse with corresponding data from the opossum – a marsupial that shares with mammals like mice or humans a common evolutionary ancestor dating back nearly 160 million years. According to Prof. Kaessmann, this comparison revealed a temporal pattern that is shared between different cell types. “The gene regulatory programmes of each cell type deviate more between species as development progresses,” explains Ioannis Sarropoulos, a doctoral researcher in Henrik Kaessmann’s research group.
The researchers explain that, at the level of individual cell types, these findings lend further support to a groundbreaking hypothesis on embryonic development from the 19th century: the Baltic German naturalist Karl Ernst von Baer (1792 to 1876) observed that the embryos of different types of vertebrates were more difficult to distinguish from one another the younger they were, thus pointing to progressive divergence in the development of vertebrate embryos.
The research results were published in the journal “Science”. In addition to the Heidelberg scientists from the ZMBH and the KiTZ, a joint institution of the German Cancer Research Center, Heidelberg University Hospital and Heidelberg University, researchers from the Museum für Naturkunde in Berlin, the Francis Crick Institute in London (Great Britain), and the Centre for Genomic Regulation in Barcelona (Spain) also participated. The European Research Council funded the research work. The research data are available in a publicly accessible database.
Contact:
Heidelberg University
Communications and Marketing
Press Office, phone +49 6221 54-2311
presse@rektorat.uni-heidelberg.de
Wissenschaftliche Ansprechpartner:
Prof. Dr Henrik Kaessmann
Center for Molecular Biology of Heidelberg University
Phone +49 6221 54-5854
h.kaessmann@uni-heidelberg.de
Originalpublikation:
I. Sarropoulos, M. Sepp, R. Frömel, K. Leiss, N. Trost, E. Leushkin, K. Okonechnikov, P. Joshi, P. Giere, L.M. Kutscher, M. Cardoso-Moreira, S.M. Pfister, and H. Kaessmann: Developmental and evolutionary dynamics of cis-regulatory elements in mouse cerebellar cells. Science (published online 29 July 2021), http://science.sciencemag.org/lookup/doi/10.1126/science.abg4696
Weitere Informationen:
http://www.zmbh.uni-heidelberg.de/Kaessmann
https://apps.kaessmannlab.org/mouse_cereb_atac
Media Contact
All latest news from the category: Life Sciences and Chemistry
Articles and reports from the Life Sciences and chemistry area deal with applied and basic research into modern biology, chemistry and human medicine.
Valuable information can be found on a range of life sciences fields including bacteriology, biochemistry, bionics, bioinformatics, biophysics, biotechnology, genetics, geobotany, human biology, marine biology, microbiology, molecular biology, cellular biology, zoology, bioinorganic chemistry, microchemistry and environmental chemistry.
Newest articles

Innovative 3D printed scaffolds offer new hope for bone healing
Researchers at the Institute for Bioengineering of Catalonia have developed novel 3D printed PLA-CaP scaffolds that promote blood vessel formation, ensuring better healing and regeneration of bone tissue. Bone is…

The surprising role of gut infection in Alzheimer’s disease
ASU- and Banner Alzheimer’s Institute-led study implicates link between a common virus and the disease, which travels from the gut to the brain and may be a target for antiviral…

Molecular gardening: New enzymes discovered for protein modification pruning
How deubiquitinases USP53 and USP54 cleave long polyubiquitin chains and how the former is linked to liver disease in children. Deubiquitinases (DUBs) are enzymes used by cells to trim protein…



