More than 100,000 switches
Graphic: Lutz Hein
Information for building cells is stored in our genetic material, otherwise known as DNA. It is here that you find all the blueprints for the more than 20,000 different proteins in the human body. Each and every cell requires several thousand different proteins in order to function.
If you were to roll every single protein blueprint into one, the information they contain would fit on less than two percent of all DNA. What are the remaining 98 percent of the genomes needed for? The switches that control gene activity are located here.
For the first time, a research team led by Dr. Ralf Gilsbach and Prof. Dr. Lutz Hein from the Institute of Experimental and Clinical Pharmacology and Toxicology at the University of Freiburg have mapped out the gene regulators in the DNA of human cardiac muscle cells. The findings have been published in the journal Nature Communications.
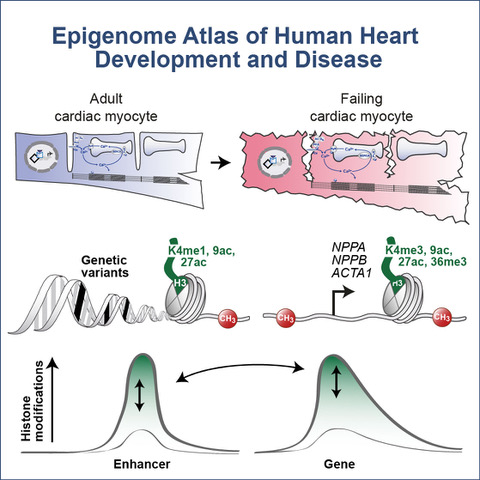
In order to locate all gene switches, the Freiburg research team used modern sequencing methods to examine the entire genome – DNA, epigenetic markers and RNA – during the development, maturation and disease of human cardiac muscle cells.
By analyzing more than a trillion sequencing letters, the team found over 100,000 gene switches. The multitude of data now yields a complete atlas of gene regulators in the life of a cardiac muscle cell. During development and growth, DNA methylation and histone markers control which genes are turned on or off.
The atlas also provides insight into mechanisms that are misdirected in heart disease. Some regulatory elements are altered in cardiac arrhythmias at the DNA level, for example. In contrast, histones take control in chronic heart failure. In the future, the Freiburg researchers want to identify the most important switches in this atlas in order to treat heart disease.
Original publication:
Ralf Gilsbach, Martin Schwaderer, Sebastian Preissl, Björn A. Grüning, David Kranzhöfer, Pedro Schneider, Thomas G. Nührenberg, Sonia Mulero-Navarro, Dieter Weichenhan, Christian Braun, Martina Dreßen, Adam R. Jacobs, Harald Lahm, Torsten Doenst, Rolf Backofen, Markus Krane, Bruce D. Gelb, Lutz Hein (2018): Distinct epigenetic programs regulate cardiac myocyte development and disease in the human heart in vivo. In: Nature Communications. http://dx.doi.org/10.1038/s41467-017-02762-z
Contact:
Prof. Dr. Lutz Hein
Institute of Experimental and Clinical Pharmacology and Toxicology
University of Freiburg
Fax: 0761/203-5314
E-Mail: Lutz.Hein@pharmakol.uni-freiburg.de
https://www.pr.uni-freiburg.de/pm-en/press-releases-2018/more-than-100-000-switc…
Media Contact
All latest news from the category: Life Sciences and Chemistry
Articles and reports from the Life Sciences and chemistry area deal with applied and basic research into modern biology, chemistry and human medicine.
Valuable information can be found on a range of life sciences fields including bacteriology, biochemistry, bionics, bioinformatics, biophysics, biotechnology, genetics, geobotany, human biology, marine biology, microbiology, molecular biology, cellular biology, zoology, bioinorganic chemistry, microchemistry and environmental chemistry.
Newest articles

First-of-its-kind study uses remote sensing to monitor plastic debris in rivers and lakes
Remote sensing creates a cost-effective solution to monitoring plastic pollution. A first-of-its-kind study from researchers at the University of Minnesota Twin Cities shows how remote sensing can help monitor and…

Laser-based artificial neuron mimics nerve cell functions at lightning speed
With a processing speed a billion times faster than nature, chip-based laser neuron could help advance AI tasks such as pattern recognition and sequence prediction. Researchers have developed a laser-based…

Optimising the processing of plastic waste
Just one look in the yellow bin reveals a colourful jumble of different types of plastic. However, the purer and more uniform plastic waste is, the easier it is to…



