NIH-Supported Researchers Map Epigenome of More Than 100 Tissue and Cell Types
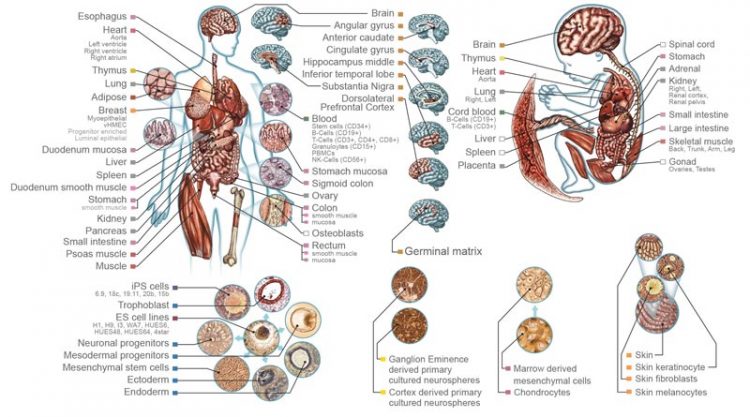
Image courtesy of Nature and Roadmap Epigenomics Consortium Reference epigenomes are available for more than 100 cell and tissue types.
Researchers supported by the National Institutes of Health Common Fund’s Roadmap Epigenomics Program ( http://commonfund.nih.gov/epigenomics/index ) have mapped the epigenomes of more than 100 types of cells and tissues, providing new insight into which parts of the genome are used to make a particular type of cell. The data, available to the biomedical research community, can be found at the National Center for Biotechnology Information website ( http://www.ncbi.nlm.nih.gov/ ).
“This represents a major advance in the ongoing effort to understand how the 3 billion letters of an individual’s DNA instruction book are able to instruct vastly different molecular activities, depending on the cellular context,” said NIH Director Francis Collins, M.D., Ph.D. “This outpouring of data-rich publications, produced by a remarkable team of creative scientists, provides powerful momentum for the rapidly growing field of epigenomics.”
Researchers from the NIH Common Fund’s Roadmap Epigenomics Program published a description of the epigenome maps in the journal Nature. More than 20 additional papers, published in Nature and Nature-associated journals, show how these maps can be used to study human biology.
“What the Roadmap Epigenomics Program has delivered is a way to look at the human genome in its living, breathing nature from cell type to cell type,” said Manolis Kellis, Ph.D., professor of computer science at the Massachusetts Institute of Technology, Cambridge, and senior author of the paper.
Understanding epigenomics
Almost all human cells have identical genomes that contain instructions on how to make the many different cells and tissues in the body. During the development of different types of cells, regulatory proteins turn genes on and off and, in doing so, establish a layer of chemical signatures that make up the epigenome of each cell. In the Roadmap Epigenomics Program, researchers compared these epigenomic signatures and established their differences across a variety of cell types. The resulting information can help us understand how changes to the genome and epigenome can lead to conditions such as Alzheimer’s disease, cancer, asthma, and fetal growth abnormalities.
The value of epigenomic data
Researchers can now take data from different cell types and directly compare them. “Today, sequencing the human genome can be done rapidly and cheaply, but interpreting the genome remains a challenge,” said Bing Ren, Ph.D., professor of cellular and molecular medicine at the University of California, San Diego, and co-author of the Nature paper and several of the associated papers. “These 111 reference epigenome maps are essentially a vocabulary book that helps us decipher each DNA segment in distinct cell and tissue types. These maps are like snapshots of the human genome in action.”
“This is the most comprehensive catalog of epigenomic data from primary human cells and tissues to date,” said Lisa Helbling Chadwick, Ph.D., project team leader and a program director at the National Institute of Environmental Health Sciences (NIEHS), part of NIH. “This coordinated effort, along with uniform data processing, makes it much easier for researchers to make direct comparisons across the entire data set.”
“Researchers from the 88 projects supported by the program, including those from this recent series of papers, have propelled the development of new epigenomic technologies,” said John Satterlee, Ph.D., co-coordinator of the Roadmap Epigenomics Program, and program director at the National Institute on Drug Abuse (NIDA), part of NIH. Satterlee added that the work of this program has served as a foundation for continued exploration of the human epigenome through the International Human Epigenome Consortium ( http://www.ihec-epigenomes.org/ ).
“With this increased understanding of the full epigenome, and the datasets available to the entire scientific community, the NIH Common Fund is striving to catalyze future research, to aid the understanding of how epigenomics plays a role in human diseases, with the expectation that further studies will identify early indications of disease and targets for therapeutics,” said James Anderson, M.D., Ph.D., director of NIH Division of Program Coordination, Planning, and Strategic Initiatives that oversees the NIH Common Fund.
NIDA, NIEHS, and the National Institute on Deafness and Other Communication Disorders are co-administrators of the NIH Common Fund’s Epigenomics Program.
Reference:
Roadmap Epigenomics Consortium. 2015. Integrative analysis of 111 reference human epigenomes. Nature; doi: 10.1038/nature14248.
The NIH Common Fund supports a series of exceptionally high impact research programs that are broadly relevant to health and disease. Common Fund programs are designed to overcome major research barriers and pursue emerging opportunities for the benefit of the biomedical research community at large. The research products of the Common Fund programs are expected to catalyze disease-specific research supported by the NIH Institutes and Centers. To learn more about the NIH Common Fund, visit http://commonfund.nih.gov .
NIDA supports most of the world’s research on the health aspects of drug abuse and addiction. The institute carries out a large variety of programs to inform policy and improve practice. Fact sheets on the health effects of drugs of abuse and information on NIDA research and other activities can be found at http://www.drugabuse.gov, which is now compatible with your smartphone, iPad, or tablet. To order publications in English or Spanish, call NIDA’s DrugPubs research dissemination center at 1-877-NIDA-NIH or 240-645-0228 (TDD) or email requests to drugpubs@nida.nih.gov. Online ordering is available at http://drugpubs.drugabuse.gov . NIDA’s media guide can be found at http://drugabuse.gov/mediaguide/ , and its easy-to-read website can be found at http://www.easyread.drugabuse.gov
NIEHS supports research to understand the effects of the environment on human health and is part of NIH. For more information on environmental health topics, visit http://www.niehs.nih.gov. Subscribe to one or more of the NIEHS news lists ( http://www.niehs.nih.gov/news/newslist/index.cfm ) to stay current on NIEHS news, press releases, grant opportunities, training, events, and publications.
About the National Institutes of Health (NIH): NIH, the nation's medical research agency, includes 27 Institutes and Centers and is a component of the U.S. Department of Health and Human Services. NIH is the primary federal agency conducting and supporting basic, clinical, and translational medical research, and is investigating the causes, treatments, and cures for both common and rare diseases. For more information about NIH and its programs, visit http://www.nih.gov
NIH…Turning Discovery Into Health®
Contacts:
Joe Balintfy, NIEHS
919-541-1993
balintfy@niehs.nih.gov
Robin Mackar, NIEHS
919-541-0073
rmackar@niehs.nih.gov
NIDA Press Office
301-443-6245
media@nida.nih.gov
Robin Mackar
News Director
rmackar@mail.nih.gov
Phone: 919-541-0073
Media Contact
All latest news from the category: Life Sciences and Chemistry
Articles and reports from the Life Sciences and chemistry area deal with applied and basic research into modern biology, chemistry and human medicine.
Valuable information can be found on a range of life sciences fields including bacteriology, biochemistry, bionics, bioinformatics, biophysics, biotechnology, genetics, geobotany, human biology, marine biology, microbiology, molecular biology, cellular biology, zoology, bioinorganic chemistry, microchemistry and environmental chemistry.
Newest articles

First-of-its-kind study uses remote sensing to monitor plastic debris in rivers and lakes
Remote sensing creates a cost-effective solution to monitoring plastic pollution. A first-of-its-kind study from researchers at the University of Minnesota Twin Cities shows how remote sensing can help monitor and…

Laser-based artificial neuron mimics nerve cell functions at lightning speed
With a processing speed a billion times faster than nature, chip-based laser neuron could help advance AI tasks such as pattern recognition and sequence prediction. Researchers have developed a laser-based…

Optimising the processing of plastic waste
Just one look in the yellow bin reveals a colourful jumble of different types of plastic. However, the purer and more uniform plastic waste is, the easier it is to…



