Protein Pairs Make Cells Remember
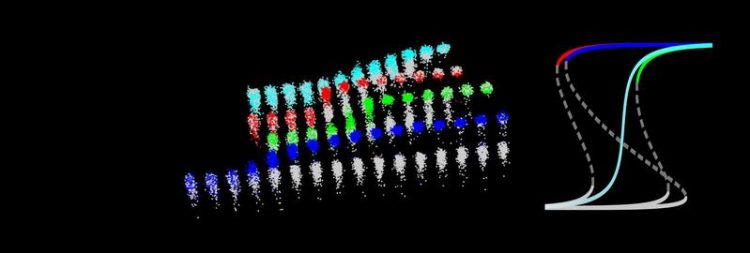
Cells with protein pairs store information for the long term (blue). Cells with single proteins do not display persistent memory (red and cyan). University of Basel, Biozentrum
Like our brains, individual cells also have a kind of memory, which enables them to store information. To make this possible, the cells require positive feedback from their proteins. The research group led by Prof. Attila Becskei at the Biozentrum of the University of Basel has now discovered that the proteins need to form pairs in these feedback loops to store information.
Cellular memory works only with protein pairs
The feedback by protein pairs works properly under specific conditions: “For dimerization the proteins must be present in the right concentration,” says Attila Becskei. If there are too few proteins, no pairs form and the cell does not store information. But when the protein concentration is too high, coupling does not work either.
“It's similar to us humans. In large cities, packed with people, dating is difficult. But living alone in the countryside does not make it easier to find a partner. So we also need to be at the right place at the right time,” illustrates Becskei.
Once the protein pairs are formed they give the cell the signal to store information in its memory. This makes the cell more sensitive to remark environmental stimuli and to respond to these more quickly in the future.
Paired protein also essential for cell differentiation
The cell not only requires the appropriate feedback from protein pairs in order to remember information but also for cell division and cell differentiation – the development of specialized cells. The understanding of the functioning of such feedback loops can reveal how to erase the cell’s memory. This is necessary, for example, for being able to turn a specialized cell, such as a skin cell, back into an unspecialized stem cell.
“For cellular reprogramming the cell must first forget that is was a skin cell,” says Becskei. “Using mathematical models we have developed, we now want to investigate, which other feedback loops contribute to cellular memory.”
Original source
Chieh Hsu, Vincent Jaquet, Mumun Gencoglu & Attila Becskei
Protein dimerization generates bistability in positive feedback loops
Cell Reports (2016), doi: 10.1016/j.celrep.2016.06.072
Further information
Prof. Dr. Attila Becskei, University of Basel, Biozentrum, tel. +41 61 267 22 22, email: attila.becskei@unibas.ch
Heike Sacher, University of Basel, Communications Biozentrum, tel. +41 61 267 14 49, email: heike.sacher@unibas.ch
Media Contact
More Information:
http://www.unibas.chAll latest news from the category: Life Sciences and Chemistry
Articles and reports from the Life Sciences and chemistry area deal with applied and basic research into modern biology, chemistry and human medicine.
Valuable information can be found on a range of life sciences fields including bacteriology, biochemistry, bionics, bioinformatics, biophysics, biotechnology, genetics, geobotany, human biology, marine biology, microbiology, molecular biology, cellular biology, zoology, bioinorganic chemistry, microchemistry and environmental chemistry.
Newest articles

First-of-its-kind study uses remote sensing to monitor plastic debris in rivers and lakes
Remote sensing creates a cost-effective solution to monitoring plastic pollution. A first-of-its-kind study from researchers at the University of Minnesota Twin Cities shows how remote sensing can help monitor and…

Laser-based artificial neuron mimics nerve cell functions at lightning speed
With a processing speed a billion times faster than nature, chip-based laser neuron could help advance AI tasks such as pattern recognition and sequence prediction. Researchers have developed a laser-based…

Optimising the processing of plastic waste
Just one look in the yellow bin reveals a colourful jumble of different types of plastic. However, the purer and more uniform plastic waste is, the easier it is to…



