Molecular Chains Line Up to Form Protopolymer
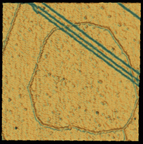
Protopolyphenylene is composed of molecules lined up for reaction and held in place by the copper substrate surface and intermolecular interactions. The image of a 270-angstrom (27-nanometer) square area of the surface was recorded with a scanning tunneling microscope at 77 K in ultrahigh vacuum. The ridged ring structure is the protopolymer; the angled lines are one-atom-high steps of the copper substrate.
First observation of extended chains of molecules that exhibit a strong interaction without forming chemical bonds
A new chemical state, designated a “protopolymer,” has been observed by Penn State researchers in chains of phenylene molecules on a crystalline copper surface at low temperature. Protopolymers form when monomers, small molecules that link together chemically to form long chains, align and interact without forming chemical bonds. The novel structures were discovered by Paul S. Weiss, professor of chemistry and physics at Penn State and Gregory S. McCarty, a graduate student at time of discovery and now a research assistant professor of engineering science and mechanics. While surface-mediated pairing and other interactions have previously been seen on metal surfaces, this is the first observation of extended chains of molecules that exhibit a strong interaction without forming chemical bonds. This type of alignment could be used to control growth and assembly of molecules and for manipulation of nanostructured materials, which are assembled on an atomic or molecular scale. Nanostructured materials often exhibit very different properties from those made by conventional techniques. A paper describing the research results, titled “Formation and Manipulation of Protopolymer Chains,” will be published in the Journal of the American Chemical Society on 15 December 2004.
Weiss points out that in substrate-mediated interactions, those in which the surface participates in the electronic interactions between molecules, the surface itself acts as a catalyst, holding molecules in place and enabling them to align for reaction. “If we use substrate-mediated interactions to direct the arrangement of monomers prior to chemical bonding, we may be able to build atomically precise structures,” says Weiss. “The key is to understand how the electronic functions of the molecule-surface interaction drive reactions and how they can be used to enhance chemical selectivity.”
The researchers carried out the experiments in a low temperature scanning tunneling microscope (STM) under ultrahigh vacuum by exposing a close-packed copper surface to p-diiodobenzene molecules. On the surface, the molecules dissociate into phenylene (cyclic C6H4) reaction intermediates and two iodine atoms. The positions of these phenylene molecules are observed by STM. Extended structures self-assembled as long chains on the surface. While individual phenylene molecules remain mobile, molecules in the chains did not move under the imaging conditions. They were able to extract molecules from the chains, however, by applying voltage pulses to the STM tip. This suggests that the chains are not covalently bound together, but instead are held by electronic reactions between molecules that are mediated by the surface. These chains extended across atomic steps on the copper surface where the level of the surface drops by one atom, resembling a stair step. This is a region in which electronic perturbations would be expected to disrupt the continuity. “It amazed us that these extended structures could cross step boundaries,” Weiss says. “These monomers have not yet formed covalent chemical bonds, which would link them together as a large molecule, but they are aligned and their interaction is much stronger than any previously observed.”
Weiss and McCarty were surprised to find that although the protopolymer is ’ready’ to form a molecule, individual units can still be manipulated and even pulled out of the chain. The protopolymer chains were stationary on the copper surface, but short chains on a phenylene-coated copper surface could be moved with the STM tip. The existence of this bonding state could potentially have significant implications for supramolecular design. These intermolecular interactions could be used to place compounds together like a jigsaw puzzle into complex structures based on the choice of assembly units and substrate surfaces–one more step toward the molecular design and engineering of new nanostructured materials.
This research was funded, in part, by the National Science Foundation, the Defense Advanced Research Projects Agency (DARPA), and the Office of Naval Research.
Media Contact
More Information:
http://www.psu.eduAll latest news from the category: Life Sciences and Chemistry
Articles and reports from the Life Sciences and chemistry area deal with applied and basic research into modern biology, chemistry and human medicine.
Valuable information can be found on a range of life sciences fields including bacteriology, biochemistry, bionics, bioinformatics, biophysics, biotechnology, genetics, geobotany, human biology, marine biology, microbiology, molecular biology, cellular biology, zoology, bioinorganic chemistry, microchemistry and environmental chemistry.
Newest articles

Innovative 3D printed scaffolds offer new hope for bone healing
Researchers at the Institute for Bioengineering of Catalonia have developed novel 3D printed PLA-CaP scaffolds that promote blood vessel formation, ensuring better healing and regeneration of bone tissue. Bone is…

The surprising role of gut infection in Alzheimer’s disease
ASU- and Banner Alzheimer’s Institute-led study implicates link between a common virus and the disease, which travels from the gut to the brain and may be a target for antiviral…

Molecular gardening: New enzymes discovered for protein modification pruning
How deubiquitinases USP53 and USP54 cleave long polyubiquitin chains and how the former is linked to liver disease in children. Deubiquitinases (DUBs) are enzymes used by cells to trim protein…



