Patient outcomes linked to biomarker levels by quantitative technology
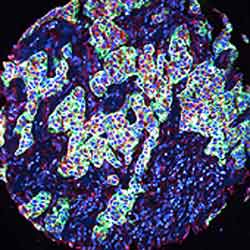
A multiplex image analyzed by AQUA to quantitatively analyze protein expression. Different colors represent compartments where protein concentration is measured; the number of pixels of each color is compared to the total number of pixels to give protein concentration.
Researchers in the Department of Pathology at Yale University School of Medicine report that when using current pathology methods of biomarker detection, the concentration of antibodies used dramatically alters the apparent relationship of biomarker level to clinical outcome. The paper appears in the December issue of the Journal of the National Cancer Institute.
The study, led by David L. Rimm, M.D., associate professor of pathology and member of the Yale Cancer Center, was designed to make sense of inconsistencies in traditional immunohistochemistry, a technique widely used for evaluating biomarker levels. The researchers evaluated levels of the common breast cancer biomarkers HER2, p53 and estrogen receptor (ER) to determine the importance of antibody standardization. They used a tissue microarray format containing specimens from 250 breast cancer patients with available long-term survival data.
“We found that the antibody concentration chosen by pathologists can dramatically affect and even reverse the apparent relationships between biomarker expression levels and patient outcomes,” said Rimm. “This work challenges the way pathologists have viewed immunohistochemistry, and points out that biomarker expression studies need further development and analysis.”
To standardize measurements, the study used the Automated Quantitative Analysis (AQUA™) technology, a system originally developed to assess biomarker expression in tissue sections. It combined fluorescence-based imaging with automated microscopy and high-throughput tissue microarray analysis technologies.
Their results with AQUA™ showed that when a high antibody concentration was used in the assay, low HER2 marker levels were associated with decreased survival. However, if a low antibody concentration was used, high HER2 levels were associated with decreased survival. Results for p53 were similar to those for HER2 but, increased ER levels were associated with increased survival regardless of antibody concentration.
These results suggest that an antibody concentration arbitrarily chosen by the pathologist may not be adequate for the expression range of the biomarker being tested. The apparent reversal of the relationship between marker level and survival rate occurs when there is a non-linear relationship between the antibody concentration and its target. ER was predicted consistently due to a linear relationship.
The findings are of importance as the relationship between protein expression and disease has an increasing role in patient treatments and individualized therapies. Variations in biomarker levels and patient response or outcome have been previously highlighted in the literature, but until now researchers have had no effective method to standardize detection techniques. Creating a new quantitative level of standardization may improve current pathology methods and practices.
“Biomarkers may have the power to provide diagnostic, therapeutic, and prognostic information for personalized medicine.” said Donald Earl Henson, M.D., of the George Washington University Cancer Institute, in “Back to the Drawing Board on Immunohistochemistry and Predictive Factors,” an accompanying editorial. “However, immunohistochemistry, a popular technique for evaluating biomarker expression, may contain procedural flaws that jeopardize its promise.”
In follow up to the editorial, Dennis C. Sgroi, M.D., director of Breast Pathology at the Massachusetts General Hospital and an associate professor of Pathology at the Harvard Medical School noted, “AQUA has proven to be a very effective tool in determining the flaws and inconsistencies associated with immunohistochemistry. These findings warrant review by pathologists in questioning the validity of current practices and before the data from predictive biomarker studies is formally integrated into practice and patient treatment.”
Co-authors at Yale include Anthony McCabe, Marisa Dolled-Filhart, Robert L. Camp. The study was funded by grants from The Department of Defense Breast Cancer Research Program, the National Cancer Institute of the National Institutes of Health, the Greenwich Breast Cancer Alliance, and the Patrick and Catherine Weldon Donaghue Foundation for Medical Research.
Camp and Rimm are founders, stockholders, and consultants to HistoRx, a private corporation to which Yale University has given exclusive rights to produce and distribute the software and technologies embedded in AQUA™. For further information contact Robert A. Curtis, bcurtis@historx.com, or 203-498-7500 or Courtney Harris courtney.harris@fkhealth.com, 617-761-6744
Citation: J. National Cancer Inst. : (December 21, 2005)
Media Contact
More Information:
http://www.yale.eduAll latest news from the category: Life Sciences and Chemistry
Articles and reports from the Life Sciences and chemistry area deal with applied and basic research into modern biology, chemistry and human medicine.
Valuable information can be found on a range of life sciences fields including bacteriology, biochemistry, bionics, bioinformatics, biophysics, biotechnology, genetics, geobotany, human biology, marine biology, microbiology, molecular biology, cellular biology, zoology, bioinorganic chemistry, microchemistry and environmental chemistry.
Newest articles

First-of-its-kind study uses remote sensing to monitor plastic debris in rivers and lakes
Remote sensing creates a cost-effective solution to monitoring plastic pollution. A first-of-its-kind study from researchers at the University of Minnesota Twin Cities shows how remote sensing can help monitor and…

Laser-based artificial neuron mimics nerve cell functions at lightning speed
With a processing speed a billion times faster than nature, chip-based laser neuron could help advance AI tasks such as pattern recognition and sequence prediction. Researchers have developed a laser-based…

Optimising the processing of plastic waste
Just one look in the yellow bin reveals a colourful jumble of different types of plastic. However, the purer and more uniform plastic waste is, the easier it is to…



