Second level of information on genome
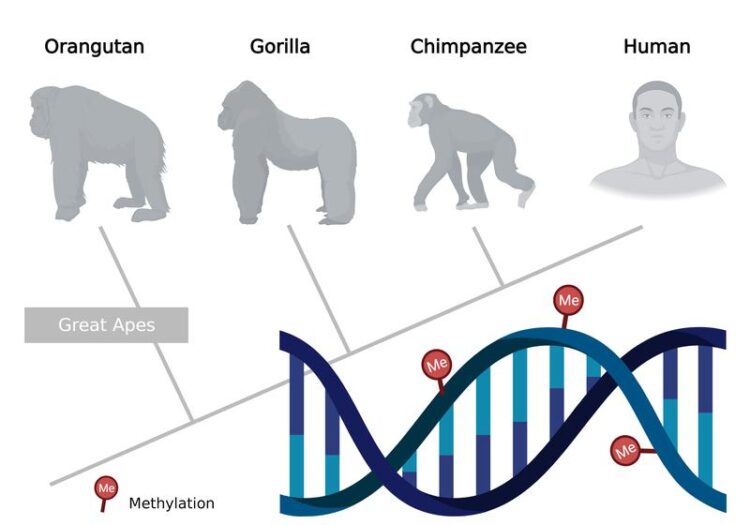
Epigenetic changes in DNA, e.g. through methylation, can be used to reconstruct phylogenetic trees.
Graphic: FLI / Kerstin Wagner; created with www.Biorender.com
Epigenetic data have enormous potential…
The cells of our body all have the same genetic material. Their genome is identical, but the cells are different, because their genes are differently active. Chemical marks on the DNA, for example through methylation or modification of histones, control the activity of the genes and shape their cell identity (epigenetics). Using methylation data from great apes, researchers at the Leibniz Institute on Aging – Fritz Lipmann Institute (FLI) in Jena, Germany, were able to show that classical sequence-based methods of phylogenetics can also be applied to the field of epigenomics. Phylogenetic trees can be correctly reconstructed and relationships explained from them.
Our genetic information is stored in DNA as a sequence of specific base pairings. This is passed on largely unchanged from one generation to the next. But how is it possible that cells differ so massively in form and function, such as in the blood, nerves, skin or teeth, even though they have all the same genetic material (genome)? This is where “epigenetics” comes into play. This primarily refers to chemical changes in our genome that ultimately influence whether and how a cell can access its genetic material. In other words, epigenetic mechanisms determine when which gene is active or inactive. Among the most important epigenetic changes are DNA methylation and the alteration of specific genome-associated proteins (histones). Such changes are also referred to as the epigenome. In contrast to the genome, the epigenome can be rapidly adjusted in response to changes (e.g., environmental factors). Research on the epigenome therefore focuses on the mechanisms that control the activity of genes without altering the base sequence of DNA.
Although the epigenome is attracting increasing attention little is known about how epigenetic changes over time. There is also a lack of systematic investigation strategies. In collaboration with human geneticist Prof. Steve Horvath from the University of California, Los Angeles, USA, researchers from the Jena Leibniz Institute on Aging – Fritz Lipmann Institute (FLI) from the research group “Computational Biology of Aging” and the Core Facility Life Science Computing have shown that classical phylogenetic methods can also be systematically applied to epigenetic data.
Phylogenetic trees based on epigenetic data
In biology, phylogenetic trees have been reconstructed based on sequence-based methods for more than 50 years to comprehensively describe evolutionary events and relationships. “When using epigenetic data, we asked ourselves whether the information they contain enables meaningful evolutionary analysis. Are the signals at all well enough conserved over a longer evolutionary distance to be able to correctly infer phylogenetic relationships from them,” says Dr. Arne Sahm, explaining the research approach.
“To gain insights into the function of individual genes and their metabolic pathways, we have used various sequence-based methods in the past, which we have now systematically transferred to the level of DNA methylation,” adds Dr. Sahm, first author of the study published in “Molecular Biology and Evolution”. DNA methylation is a direct chemical modification of DNA bases carried out by specialized enzymes in a complicated process. Changes in the start region of genes are particularly consequential: methylation can directly control the activity of a gene.
Epigenetics enables more reliable reconstruction of ancestral relationships
Since our closest relatives – chimpanzees, gorillas and orangutans – differ only slightly from humans in their genomes, the researchers used a dataset of blood samples from great apes for their analyses. “We were able to show that reconstructing phylogenetic trees based on DNA methylation data works very well,” reports Prof. Steve Hoffmann, research group leader at FLI and professor of computational biology at Friedrich Schiller University in Jena, Germany. “We were particularly surprised that the ancestral relationships of the great apes can be reconstructed even more reliably with the epigenomic data than with the genomic data. Since methylation is affected by environmental factors or disease, this was not expected.”
DNA methylation strongly conserved
The FLI researchers were also able to show that DNA methylation differs only very slightly among the great ape species studied. Thus, genome segments that are highly methylated in humans are generally also altered in the great apes. The same is true for parts of the genome that are poorly methylated or not methylated at all. It is obvious that these epigenomic patterns have remained largely unchanged over millions of years of evolution, i.e., they are strongly conserved. “Here, the resolution of conservation is so high that two equivalent DNA building blocks of different great ape species are on average more similar in their methylation than two directly neighboring DNA building blocks of the same species,” Prof. Hoffmann points out.
Insights into tissue-specific phylogenetic information
Unlike the genome, which is nearly identical in all cells of an individual, the epigenome can reflect tissue-specific phylogenetic information. Therefore, comparisons between different species provide the opportunity to gain insight into the evolution of tissues, not just the whole organism. The inclusion of other epigenetic data, such as histone modifications, may be helpful to better understand such processes in the future.
Potential for deeper insights into evolutionary differences and similarities
“The application and further improvement of our developed methods promises new insights into the mechanisms of epigenetic gene regulation and the resulting emergence of new phenotypes,” explains Dr. Sahm. The methods could help to identify those parts of the genome that are most likely to play an important role in gene regulation because they have changed little over millions of years. “Conversely, we can also use the method described to identify sections of the epigenome that have changed particularly rapidly in certain species during evolution to study how these epigenetic changes influence the emergence of phenotypes.”
Publication
An analysis of methylome evolution in primates. Sahm A, Koch P, Horvath S, Hoffmann S. Mol Biol Evol. 2021, 38(11), 4700-4714. doi: 10.1093/molbev/msab189.
https://academic.oup.com/mbe/article/38/11/4700/6310173
Contact
Dr. Kerstin Wagner
Press and Public Relations
Phone: 03641-656378, email: presse@leibniz-fli.de
Background information
The Leibniz Institute on Aging – Fritz Lipmann Institute (FLI) – upon its inauguration in 2004 – was the first German research organization dedicated to research on the process of aging. More than 350 employees from around 40 nations explore the molecular mechanisms underlying aging processes and age-associated diseases. For more information, please visit http://www.leibniz-fli.de.
The Leibniz Association connects 97 independent research institutions that range in focus from natural, engineering and environmental sciences to economics, spatial and social sciences and the humanities. Leibniz Institutes address issues of social, economic and ecological relevance. They conduct basic and applied research, including in the interdisciplinary Leibniz Research Alliances, maintain scientific infrastructure, and provide research-based services. The Leibniz Association identifies focus areas for knowledge transfer, particularly with the Leibniz research museums. It advises and informs policymakers, science, industry and the general public. Leibniz institutions collaborate intensively with universities – including in the form of Leibniz ScienceCampi – as well as with industry and other partners at home and abroad. They are subject to a transparent, independent evaluation procedure. Because of their importance for the country as a whole, the Leibniz Association Institutes are funded jointly by Germany’s central and regional governments. The Leibniz Institutes employ around 20,500 people, including 11,500 researchers. The financial volume amounts to 2 billion euros. For more information: http://www.leibniz-gemeinschaft.de/en/.
Originalpublikation:
An analysis of methylome evolution in primates. Sahm A, Koch P, Horvath S, Hoffmann S. Mol Biol Evol. 2021, 38(11), 4700-4714. doi: 10.1093/molbev/msab189.
https://academic.oup.com/mbe/article/38/11/4700/6310173
Media Contact
All latest news from the category: Life Sciences and Chemistry
Articles and reports from the Life Sciences and chemistry area deal with applied and basic research into modern biology, chemistry and human medicine.
Valuable information can be found on a range of life sciences fields including bacteriology, biochemistry, bionics, bioinformatics, biophysics, biotechnology, genetics, geobotany, human biology, marine biology, microbiology, molecular biology, cellular biology, zoology, bioinorganic chemistry, microchemistry and environmental chemistry.
Newest articles

Innovative 3D printed scaffolds offer new hope for bone healing
Researchers at the Institute for Bioengineering of Catalonia have developed novel 3D printed PLA-CaP scaffolds that promote blood vessel formation, ensuring better healing and regeneration of bone tissue. Bone is…

The surprising role of gut infection in Alzheimer’s disease
ASU- and Banner Alzheimer’s Institute-led study implicates link between a common virus and the disease, which travels from the gut to the brain and may be a target for antiviral…

Molecular gardening: New enzymes discovered for protein modification pruning
How deubiquitinases USP53 and USP54 cleave long polyubiquitin chains and how the former is linked to liver disease in children. Deubiquitinases (DUBs) are enzymes used by cells to trim protein…



