Single-cell RNA seq method developed to accurately quantify cell-specific drug effects in pancreatic islets
Graphical abstract B. Marquina-Sánchez N. Fortelny
The pancreas is an abdominal organ that produces digestive enzymes as well as hormones that regulate blood sugar levels. This hormone-producing function is localized to the islets of Langerhans, which constitute clusters of different endocrine cell types.
Among those are beta cells, which produce the hormone insulin needed to lower glucose (a type of sugar) levels in our blood, as well as alpha cells, which generate the hormone glucagon in charge of raising glucose levels in the blood.
Type 1 diabetes is a chronic disease in which the body’s immune system mistakenly attacks and destroys the pancreas’ insulin-producing beta cells. Regenerative medicine aims to replenish beta cell mass, and thus support and ultimately substitute the current insulin replacement therapies.
Alterations to islet composition, including insufficient beta cell function and beta cell dedifferention, also contribute to type II diabetes. Therefore, a deeper understanding of the identity and crosstalk of the different islet cell types leads to a better characterization of both forms of diabetes and may contribute to the development of novel therapeutic concepts.
Single-cell transcriptomics is a powerful technique to characterize cellular identity. Previously, CeMM researchers from Christoph Bock’s and Stefan Kubicek’s groups at CeMM published the first single cell transcriptomes from primary human pancreatic islet cells (EMBO Rep. 2016 Feb 17;(2):178-87. DOI: 10.15252/embr.201540946).
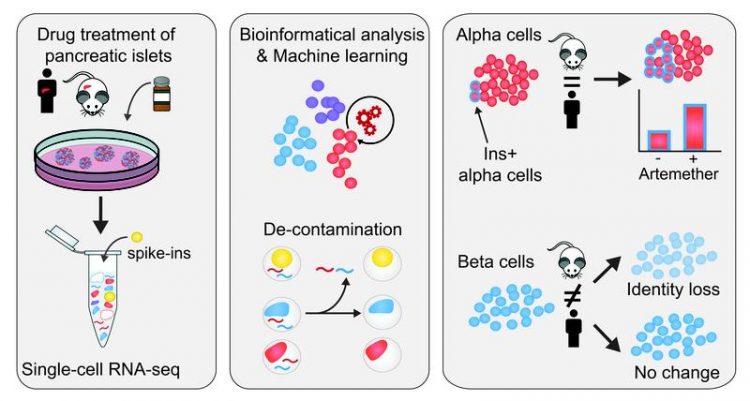
Advances in the technology have since enabled its application to the generation of global human and mouse single cell transcriptome atlases. Despite these advances, single cell approaches remain technologically challenging given that the miniscule RNA amount present is entirely used up in the experiment. Therefore, it is essential to ensure the quality and purity of the resulting single cell transcriptomes.
CeMM researchers in the two contributing laboratories identified unexpectedly high hormone expression in non-endocrine cell types, both in their own dataset as well as other published single cell studies. They set out to elucidate whether this would be the result of contamination by RNA molecules, for example from dying cells, and how it could be removed to obtain a more reliable dataset.
Such contamination seems present in single cell RNA-seq data from most tissues but was most visible in pancreatic islets. Islet endocrine cells are exclusively devoted to the production of single hormones, and insulin in beta cells and glucagon in alpha cells are expressed to higher levels than typical “housekeeping” genes. Thus, redistribution of these transcripts to other cell types was highly pronounced. Based on this observation, their goal was to develop, validate and apply a method to experimentally determine and computationally remove such contamination.
In their investigation, CeMM researchers used spiked-in cells from different cell types, both mouse and human samples, that they added to their pancreatic islet samples. Importantly, the transcriptomes of these spike-in cell were fully characterized.
This allowed them to control internally and accurately the level of RNA contamination in single cell RNA-seq, giving that the human transcripts detected in the mouse spike-in cells constitute contaminating RNA.
In this way, they found that the samples had a contamination level of up to 20%, and were able to define the contamination in each samples. They then developed a novel bioinformatics approach to computationally remove contaminating reads from single cell transcriptomes.
Having now obtained a “decontaminated” transcriptome, from which the spurious signal has been removed, they proceeded to characterize how the cellular identity in the different cell types responded to the treatment with three different drugs. They found that a small molecule inhibitor of the transcription factor FOXO1 induces dedifferentiation of both alpha and beta cells.
Furthermore, they studied artemether, which had been found to diminish the function of alpha cells and could induce insulin production in both in vivo and in vitro studies (Cell. 2017 Jan 12;168(1-2):86-100.e15. DOI: 10.1016/j.cell.2016.11.010). The effects of the drug artemether were species-specific and cell-type-specific.
In alpha cells, a fraction of cells increases insulin expression and gain aspects of beta cell identity, both in mouse and human samples. Importantly, researchers found that in human beta cells, there is no significant change in insulin expression, whereas in mouse islets, beta cells reduce their insulin expression and overall beta cell identity.
This study is the result of a cross-disciplinary collaboration of the laboratories of Stefan Kubicek and Christoph Bock at CeMM with Patrick Collombat at the Institute of Biology Valrose (France). This is the first study to apply single cell sequencing to analyze dynamic drug response in intact isolated tissue, which benefitted from the high quantitative accuracy of the decontamination method.
It provides thus not only a novel method for single-cell decontamination and highly quantitative single-cell analysis of drug responses in intact tissues, but also addresses an important current question in islet cell biology and diabetes research. These findings could open up potential therapeutic avenues to treat Type 1 diabetes in the future.
The study “Single-cell RNA-seq with spike-in cells enables accurate quantification of cell-specific drug effects in pancreatic islets” was published in Genome Biology on 6 May 2020. DOI: 10.1186/s13059-020-02006-2
Authors:
Brenda Marquina-Sanchez*, Nikolaus Fortelny*, Matthias Farlik*, Andhira Vieira, Patrick Collombat, Christoph Bock, Stefan Kubicek * shared first authorships
Funding:
The study was funded by the Juvenile Diabetes Research Foundation (JDRF SRA 201304452). Research in the Kubicek lab is also supported by the Austrian Academy of Sciences, the Austrian Federal Ministry for Digital and Economic Affairs and the National Foundation for Research, Technology, and Development, the Austrian Science Fund (FWF Special Research Program F4701-614 B20) and the European Research Council (ERC) under the European Union’s Horizon research and innovation programme (ERC-CoG-772437). Brenda Marquina-Sánchez is supported by a Boehringer Ingelheim Fonds PhD fellowship.
Christoph Bock and his lab are supported by a New Frontiers Group award of the Austrian Academy of Sciences and by an ERC Starting Grant under the European Union’s Horizon 2020 research and innovation programme (ERC-StG-679146). Nikolas Fortelny is supported by an Innovation Fund of the Austrian Academy of Sciences (IF_2015_36) and the Austrian Science Fund (FWF Special Research Program SFB-F61.02).
Stefan Kubicek joined CeMM in August 2010. He obtained an MSc in synthetic organic chemistry from the Vienna University of Technology after writing a diploma thesis at ETH Zurich. For his PhD in Thomas Jenuwein’s lab at the IMP in Vienna, he changed fields to molecular biology. He then performed postdoctoral research working on chemical biology with Stuart Schreiber at the Broad Institute of Harvard and MIT.
Stefan Kubicek heads the chemical screening platform and PLACEBO (Platform Austria for Chemical Biology), a task he is well equipped for based on previous screening experience with Boehringer Ingelheim and at the Broad Institute. Stefan Kubicek has also headed the Christian Doppler Laboratory for Chemical Epigenetics and Antiinfectives, a public-private partnership between CeMM, Boehringer Ingelheim and Haplogen. The Kubicek lab is working on the role of chromatin in the definition of cell types and cell states, particularly chromatin modifying enzymes as synthetic lethal targets in cancer and chemical transdifferentiation to insulin-producing beta cells. In an ERC-funded project, the laboratory is working on metabolic enzymes in the cell’s nucleus and testing the hypothesis that small molecule metabolites shape chromatin structure and thus control gene expression and cell identity.
Christoph Bock joined CeMM as Principal Investigator in 2012. He pursues interdisciplinary research aimed at understanding the epigenetic and gene-regulatory basis of cancer, and advancing precision medicine with genomics technology. His research group combines experimental biology (high-throughput sequencing, epigenetics, CRISPR screening, synthetic biology) with computer science (bioinformatics, machine learning, artificial intelligence). He is also a guest professor at the Medical University of Vienna, scientific coordinator of the Biomedical Sequencing Facility (BSF) at CeMM, and key researcher at the Ludwig Boltzmann Institute for Rare and Undiagnosed Diseases. He coordinates an EU Horizon 2020 project on the single-cell analysis of human organoids as a contribution to the Human Cell Atlas. Christoph Bock is an elected member of the Young Academy of the Austrian Academy of Sciences and has received major research awards, including the Max Planck Society’s Otto Hahn Medal (2009), an ERC Starting Grant (2016–2021), and the Overton Prize of the International Society of Computational Biology (2017).
The mission of CeMM Research Center for Molecular Medicine of the Austrian Academy of Sciences is to achieve maximum scientific innovation in molecular medicine to improve healthcare. At CeMM, an international and creative team of scientists and medical doctors pursues free-minded basic life science research in a large and vibrant hospital environment of outstanding medical tradition and practice. CeMM’s research is based on post-genomic technologies and focuses on societally important diseases, such as immune disorders and infections, cancer and metabolic disorders. CeMM operates in a unique mode of super-cooperation, connecting biology with medicine, experiments with computation, discovery with translation, and science with society and the arts. The goal of CeMM is to pioneer the science that nurtures the precise, personalized, predictive and preventive medicine of the future. CeMM trains a modern blend of biomedical scientists and is located at the campus of the General Hospital and the Medical University of Vienna.
Stefan Kubicek
SKubicek@cemm.oeaw.ac.at
Christoph Bock
CBock@cemm.oeaw.ac.at
The study “Single-cell RNA-seq with spike-in cells enables accurate quantification of cell-specific drug effects in pancreatic islets” was published in Genome Biology on 6 May 2020. DOI: 10.1186/s13059-020-02006-2
https://cemm.at/index.php?id=35&no_cache=1&tx_news_pi1%5Bnews_preview%5D…
Media Contact
All latest news from the category: Life Sciences and Chemistry
Articles and reports from the Life Sciences and chemistry area deal with applied and basic research into modern biology, chemistry and human medicine.
Valuable information can be found on a range of life sciences fields including bacteriology, biochemistry, bionics, bioinformatics, biophysics, biotechnology, genetics, geobotany, human biology, marine biology, microbiology, molecular biology, cellular biology, zoology, bioinorganic chemistry, microchemistry and environmental chemistry.
Newest articles

First-of-its-kind study uses remote sensing to monitor plastic debris in rivers and lakes
Remote sensing creates a cost-effective solution to monitoring plastic pollution. A first-of-its-kind study from researchers at the University of Minnesota Twin Cities shows how remote sensing can help monitor and…

Laser-based artificial neuron mimics nerve cell functions at lightning speed
With a processing speed a billion times faster than nature, chip-based laser neuron could help advance AI tasks such as pattern recognition and sequence prediction. Researchers have developed a laser-based…

Optimising the processing of plastic waste
Just one look in the yellow bin reveals a colourful jumble of different types of plastic. However, the purer and more uniform plastic waste is, the easier it is to…



