Tracked step for step: ATP splitting in membrane protein dynamically measured for the first time
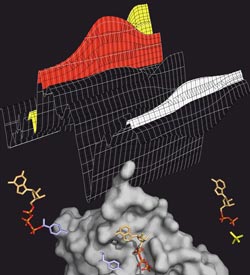
ATP splitting: The transport protein MsbA (grey) splits ATP (coloured), to generate energy for the transport process. ATP has three phosphate groups (orange-red). If one of them is split off (yellow), energy is released. The splitting process can be tracked in the infrared spectrum (above), in which the various ATP intermediate products leave characteristic bands (red: ATP, yellow: split-off phosphate, white: protein).<br>Image: Falk Syberg<br>
How a transport protein obtains its driving force from the energy storage molecule ATP, has been tracked dynamically by RUB researchers. Using time-resolved infrared spectroscopy, they measured the structural changes in the bacterial membrane protein MsbA and its interaction partner ATP.
The researchers led by Prof. Dr. Eckhard Hofmann and Prof. Dr. Klaus Gerwert from the Biophysics Department report on the results in the current issue of the Journal of Biological Chemistry.
Transport proteins are associated with various diseases
ABC transporters are membrane proteins that transport various substances from one side of the cell membrane to the other. The driving force for this is provided by the molecule ATP, a universal energy storage of the cells. ATP has three phosphate groups. If one of these splits off, energy is released. The transporters are of great medical significance as they play a central role in the multi-drug resistance of cancer cells to chemotherapeutic substances and are associated with various inherited diseases like cystic fibrosis.
In recent years, researchers have uncovered the 3D structures of several of these transporters at the atomic level. Although the architecture of the nanomachines is known, a detailed understanding of how the splitting of the energy carrier ATP dynamically enables the transport of various substances across biological membranes has so far been lacking.
Protein controls ATP splitting
The Bochum researchers have now dynamically tracked the ATP splitting, called hydrolysis, for the first time in the fat transporter MsbA from the bacterium Escherichia coli. Using fourier transform infrared spectroscopy, they studied the motor domains of MsbA, i.e. the part of the protein where the ATP splitting takes place. Using this method, researchers can track minute changes in the protein in the range of nanoseconds. Simultaneously, the method also records changes in the molecules the protein interacts with – in this case ATP.
Phosphate signals reveal what happens during the splitting
The big challenge presented by the data analysis is to assign the signals in the measured spectrum to specific molecules or molecular groups. If this is successful, you can see which groups of molecules are structurally changed and when. The biophysicists marked the phosphate groups of the ATP molecule, so that they left characteristic signals in the spectrum. In this way they tracked, how ATP bound to the transport protein, how one of its three phosphate groups split off and was released into the environment without first latching back on to the protein. “Our data also provides important clues as to how the protein moves during ATP hydrolysis. This lays the foundation for the study of the whole membrane protein, which we are going to tackle next”, says Professor Hofmann. The investigations were supported by the Protein Research Department at the RUB and funds of the collaborative research centre SFB 642 “GTP and ATP dependent membrane processes”, whose speaker is Prof. Dr. Klaus Gerwert.
Bibliographic record
F. Syberg, Y. Suveyzdis, C. Kötting, K. Gerwert, E. Hofmann (2012): Time-resolved fourier transform infrared spectroscopy of the nucleotide-binding domain from the ATP-binding cassette transporter MsbA. ATP Hydrolysis is the rate-limiting step in the catalytic cycle, Journal of Biological Chemistry, doi: 10.1074/jbc.M112.359208
Further information
Prof. Dr. Eckhard Hofmann, Protein Crystallography, Faculty of Biology and Biotechnology at the Ruhr-Universität, 44780 Bochum, Germany, tel. +49/234/32-24463, eckhard.hofmann@bph.rub.de
Click for more
Biophysics at the RUB
http://www.bph.ruhr-uni-bochum.de/index_en.htm
Editor: Dr. Julia Weiler
Media Contact
More Information:
http://www.ruhr-uni-bochum.deAll latest news from the category: Life Sciences and Chemistry
Articles and reports from the Life Sciences and chemistry area deal with applied and basic research into modern biology, chemistry and human medicine.
Valuable information can be found on a range of life sciences fields including bacteriology, biochemistry, bionics, bioinformatics, biophysics, biotechnology, genetics, geobotany, human biology, marine biology, microbiology, molecular biology, cellular biology, zoology, bioinorganic chemistry, microchemistry and environmental chemistry.
Newest articles

Innovative 3D printed scaffolds offer new hope for bone healing
Researchers at the Institute for Bioengineering of Catalonia have developed novel 3D printed PLA-CaP scaffolds that promote blood vessel formation, ensuring better healing and regeneration of bone tissue. Bone is…

The surprising role of gut infection in Alzheimer’s disease
ASU- and Banner Alzheimer’s Institute-led study implicates link between a common virus and the disease, which travels from the gut to the brain and may be a target for antiviral…

Molecular gardening: New enzymes discovered for protein modification pruning
How deubiquitinases USP53 and USP54 cleave long polyubiquitin chains and how the former is linked to liver disease in children. Deubiquitinases (DUBs) are enzymes used by cells to trim protein…



