Understanding of ocean’s role in carbon cycling
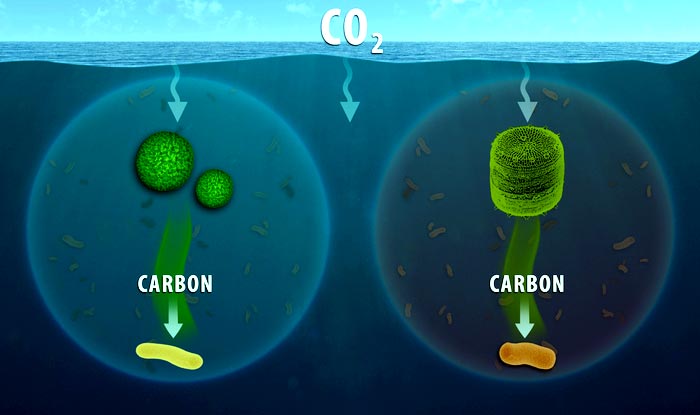
Depiction of what was studied.
Credit: Oregon State University
Microbiology researchers at Oregon State University have shed new light on the mechanisms of carbon cycling in the ocean, using a novel approach to track which microbes are consuming different types of organic carbon produced by common phytoplankton species.
The research is an important step toward forecasting how much carbon will leave the ocean for the atmosphere as greenhouse gas carbon dioxide and how much will end up entombed in marine sediments, said Ryan Mueller, associate professor in OSU’s Department of Microbiology and the leader of the study.
Findings were published today in the Proceedings of the National Academy of Sciences.
“Our research shows that different species of microbes in the ocean are very particular yet predictable in the food sources they prefer to eat,” said first author Brandon Kieft, a recent Oregon State Ph.D. graduate who is now a postdoctoral researcher at the University of British Columbia. “As global climate change continues to alter oceanic environments at a rapid pace, the availability of food sources for microbes will also change, ultimately favoring certain types over others.”
Phytoplankton are microscopic organisms at the base of the ocean’s food chain and a key component of a critical biological carbon pump. Most float in the upper part of the ocean, where sunlight can easily reach them.
The tiny autotrophic plants – they make their own food – have a big effect on the levels of carbon dioxide in the atmosphere by sucking it up during photosynthesis. It’s a natural sink and one of the primary ways that CO2, the most abundant greenhouse gas, is scrubbed from the atmosphere; atmospheric carbon dioxide has increased 40% since the dawn of the industrial age, contributing heavily to a warming planet.
“We’re studying the consumers – the heterotrophic microbes – of the organic material made by the primary producers, the microbial phytoplankton,” Mueller said. “Both groups are microbes, the former primarily consumes organic carbon as a food source, while the latter ‘fix’ their own organic carbon. Microbes form the basis of the food web and biological carbon pump, and our work is primarily focused on exploring what the consumers are doing in this system.”
The surface ocean stores nearly as much carbon as exists in the atmosphere. As the ocean pulls in atmospheric carbon dioxide, phytoplankton use the CO2 and sunlight for photosynthesis: They convert them into sugars and other compounds the cells can use for energy, producing oxygen in the process.
This so-called fixed carbon makes up the diet of heterotrophic microbes and higher organisms of the marine food web such as fish and mammals, which ultimately convert the carbon back to atmospheric CO2 through respiration or contribute to the carbon stock at the bottom of the ocean when they die and sink.
The collective respiratory activity of the heterotrophic microbial consumers is the main way that fixed dissolved organic carbon from phytoplankton is returned to the atmosphere as CO2.
Mueller, Kieft and collaborators at the Oak Ridge and Lawrence Livermore national laboratories and the universities of Tennessee, Washington and Oklahoma used stable isotope labeling to track carbon as it made its way into the organic matter produced by the phytoplankton and, ultimately, the heterotrophic microbes that consume it.
The scientists used those isotopes to tell which organisms were eating diatoms and which were consuming cyanobacteria, two species of phytoplankton that combine to produce a majority of the ocean’s fixed carbon. The researchers could also tell when the consumption was happening – for example, at times the phytoplankton cells were producing substances known as lysates during their death phase or exudates during their growth phase.
“Our findings have important implications for understanding how marine microbes and photosynthetic algae function together to impact global carbon cycling and how this oceanic food web may respond to continued environmental change,” Kieft said. “This will help us predict how much carbon will go back into the atmosphere and how much will be buried in marine sediments for centuries.”
The research was funded by the Gordon and Betty Moore Foundation Marine Biology Initiative and the U.S. Department of Energy.
Journal: Proceedings of the National Academy of Sciences
DOI: 10.1073/pnas.2101178118
Method of Research: Observational study
Subject of Research: Not applicable
Article Title: Phytoplankton exudates and lysates support distinct microbial consortia with specialized metabolic and ecophysiological traits
Article Publication Date: 7-Oct-2021
Media Contact
Steve Lundeberg
Oregon State University
steve.lundeberg@oregonstate.edu
Office: 541-737-4039
Original Source
All latest news from the category: Life Sciences and Chemistry
Articles and reports from the Life Sciences and chemistry area deal with applied and basic research into modern biology, chemistry and human medicine.
Valuable information can be found on a range of life sciences fields including bacteriology, biochemistry, bionics, bioinformatics, biophysics, biotechnology, genetics, geobotany, human biology, marine biology, microbiology, molecular biology, cellular biology, zoology, bioinorganic chemistry, microchemistry and environmental chemistry.
Newest articles

Innovative 3D printed scaffolds offer new hope for bone healing
Researchers at the Institute for Bioengineering of Catalonia have developed novel 3D printed PLA-CaP scaffolds that promote blood vessel formation, ensuring better healing and regeneration of bone tissue. Bone is…

The surprising role of gut infection in Alzheimer’s disease
ASU- and Banner Alzheimer’s Institute-led study implicates link between a common virus and the disease, which travels from the gut to the brain and may be a target for antiviral…

Molecular gardening: New enzymes discovered for protein modification pruning
How deubiquitinases USP53 and USP54 cleave long polyubiquitin chains and how the former is linked to liver disease in children. Deubiquitinases (DUBs) are enzymes used by cells to trim protein…



